EX-99.1
Published on May 30, 2017

Annual Meeting and Investor Day May 25, 2017 Exhibit 99.1

Meeting Agenda – May 25th 8:00 – 8:30am Annual Shareholders Meeting Analyst/Investor Day Presentations 8:30 – 9:15am CEO Review 9:15 – 10:00am Pathologist Perspective 10:00 – 10:15am Break 10:15 – 11:00am Pharma Services 11:00 – 12:00am R&D and New Test Development Noon - 1:00pmLunch/Review of Governmental Affairs 12:10pm – 1:30pm- Lab Tours – leaving every 20 minutes

Annual Shareholder Meeting May 25, 2017

Investor/Analyst Day Presentations May 25, 2017

Meeting Agenda – May 25th 8:00 – 8:30am Annual Shareholders Meeting Analyst/Investor Day Presentations 8:30 – 9:15am CEO Review 9:15 – 10:00am Pathologist Perspective 10:00 – 10:15am Break 10:15 – 11:00am Pharma Services 11:00 – 12:00am R&D and New Test Development Noon - 1:00pmLunch/Review of Governmental Affairs 12:10pm – 1:30pm- Lab Tours – leaving every 20 minutes

Forward-looking Statements This presentation contains statements which constitute forward-looking statements within the meaning of Section 27A of the Securities Act, as amended; Section 21E of the Securities Exchange Act of 1934; and the Private Securities Litigation Reform Act of 1995. The words “may”, “would”, “could”, “will”, “expect”, “estimate”, “anticipate”, “believe”, “intend”, “plan”, “goal”, and similar expressions and variations thereof are intended to specifically identify forward-looking statements. All statements that are not statements of historical fact are forward-looking statements. Investors and prospective investors are cautioned that any such forward-looking statements are not guarantees of future performance and involve risks and uncertainties, and that actual results may differ materially from those projected in the forward-looking statements as a result of various factors. The risks that might cause such differences are identified in our filings with the Securities and Exchange Commission. We undertake no obligation to publicly update or revise the forward looking statements made in this presentation to reflect events or circumstances after the date of this presentation or to reflect the occurrence of unanticipated events.

CEO Summary Douglas VanOort Chairman & CEO

NeoGenomics Vision By providing uncompromising quality, exceptional service and innovative solutions, we will be the World’s leading cancer testing and information company.

NeoGenomics - Facts # 1 or 2 largest somatic cancer testing Lab in America Approximately 650,000 tests will be performed in 2017 Approx. 160,000 SF of space, 1,000 employees, 50 Physicians and PhDs Most advanced and comprehensive cancer testing menu in America Outstanding franchise with Hospitals/Pathologists Unique and Growing business with Pharma clients Large market, fragmented industry, and very positive demographic trends
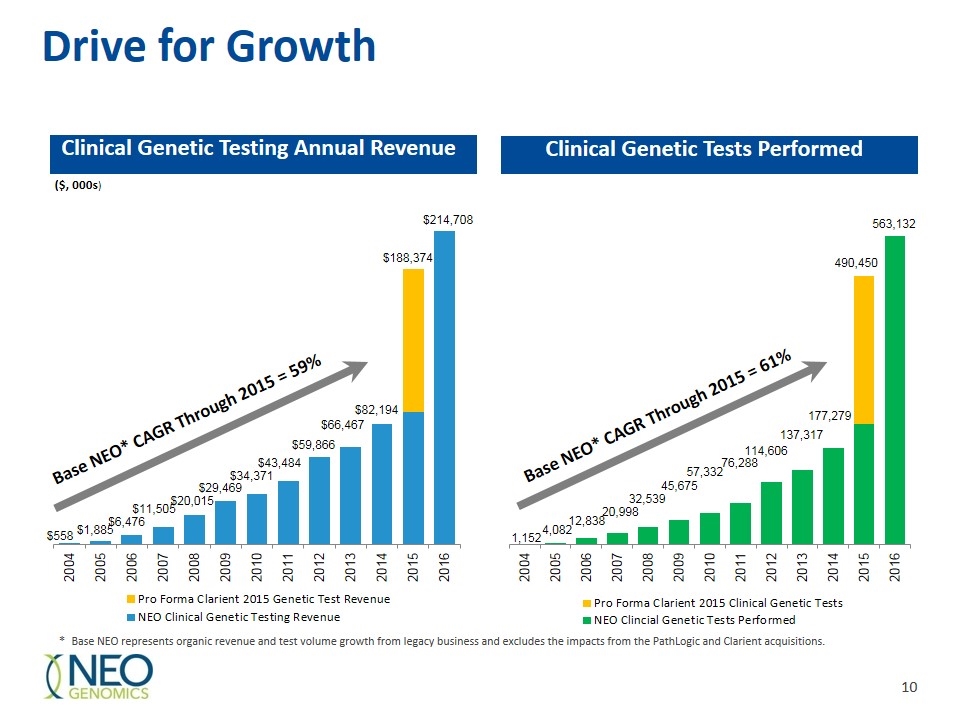
Drive for Growth Clinical Genetic Testing Annual Revenue Clinical Genetic Tests Performed ($, 000s) * Base NEO represents organic revenue and test volume growth from legacy business and excludes the impacts from the PathLogic and Clarient acquisitions. Base NEO* CAGR Through 2015 = 59% Base NEO* CAGR Through 2015 = 61%
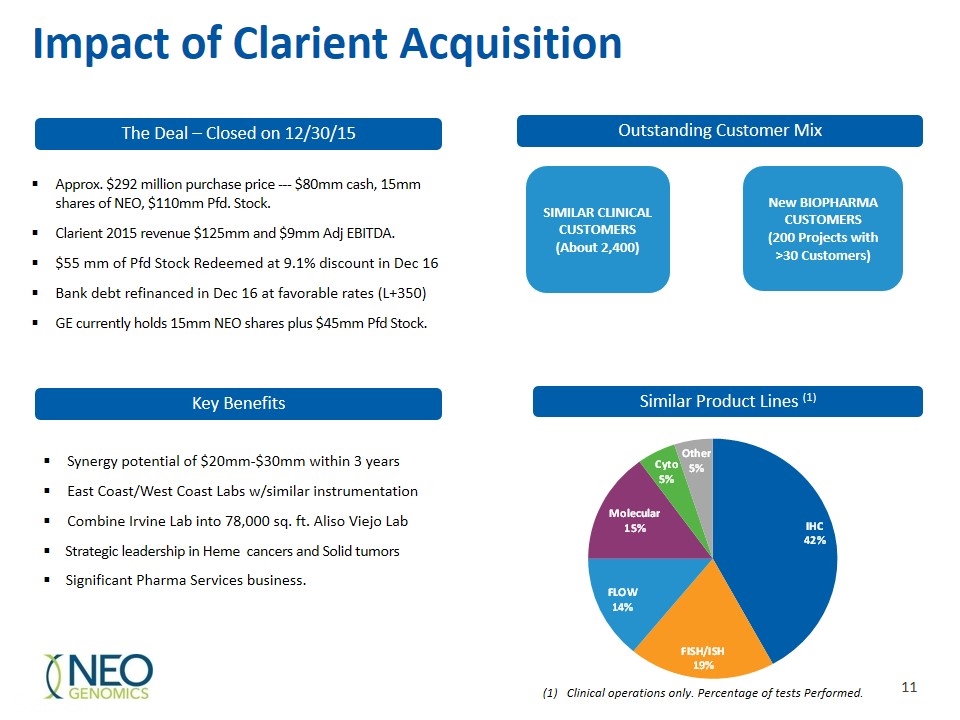
Impact of Clarient Acquisition Key Benefits Outstanding Customer Mix Similar Product Lines (1) Synergy potential of $20mm-$30mm within 3 years East Coast/West Coast Labs w/similar instrumentation Combine Irvine Lab into 78,000 sq. ft. Aliso Viejo Lab Strategic leadership in Heme cancers and Solid tumors Significant Pharma Services business. Clinical operations only. Percentage of tests Performed. SIMILAR CLINICAL CUSTOMERS (About 2,400) New BIOPHARMA CUSTOMERS (200 Projects with >30 Customers) The Deal – Closed on 12/30/15 Approx. $292 million purchase price --- $80mm cash, 15mm shares of NEO, $110mm Pfd. Stock. Clarient 2015 revenue $125mm and $9mm Adj EBITDA. $55 mm of Pfd Stock Redeemed at 9.1% discount in Dec 16 Bank debt refinanced in Dec 16 at favorable rates (L+350) GE currently holds 15mm NEO shares plus $45mm Pfd Stock.
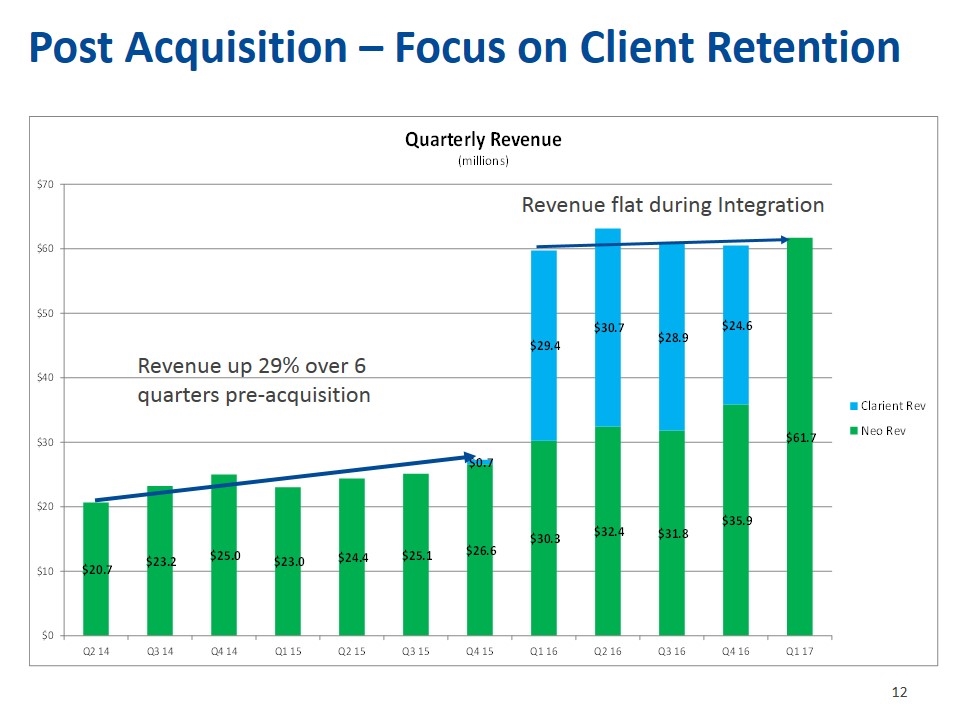
Post Acquisition – Focus on Client Retention Revenue up 29% over 6 quarters pre-acquisition Revenue flat during Integration

Integration Complete After 15 months, as of March 31, 2017, Integration is 100% complete. Sales force completely integrated after 2 months All “back - office” systems and processes integrated after 6 months LIS reprogrammed to incorporate Best Practices after 8 months Corporate culture and incentive programs fully integrated within 9-12 months All clients serviced from common LIS and Billing system after 12 months Irvine Lab completely consolidated into Aliso Viejo Lab after 15 months After 16 months, Client Retention is nearly 100%.
New NeoGenomics Same historically successful attributes : Growth orientation Leadership in Innovation Entrepreneurialism Exceptional Quality/Service Deep/comprehensive test menu Low-cost testing processes Strong culture Scale from doubling our volume Improved Profitability

Pursuit of Growth - Again Growth Goals: Mid-teens organic clinical volume growth 20%+ organic Pharma revenue growth 25-35% incremental Adjusted EBITDA on revenue growth Driven by: Market share gains Demographics and medical advances Innovation in test development Increased efficiencies
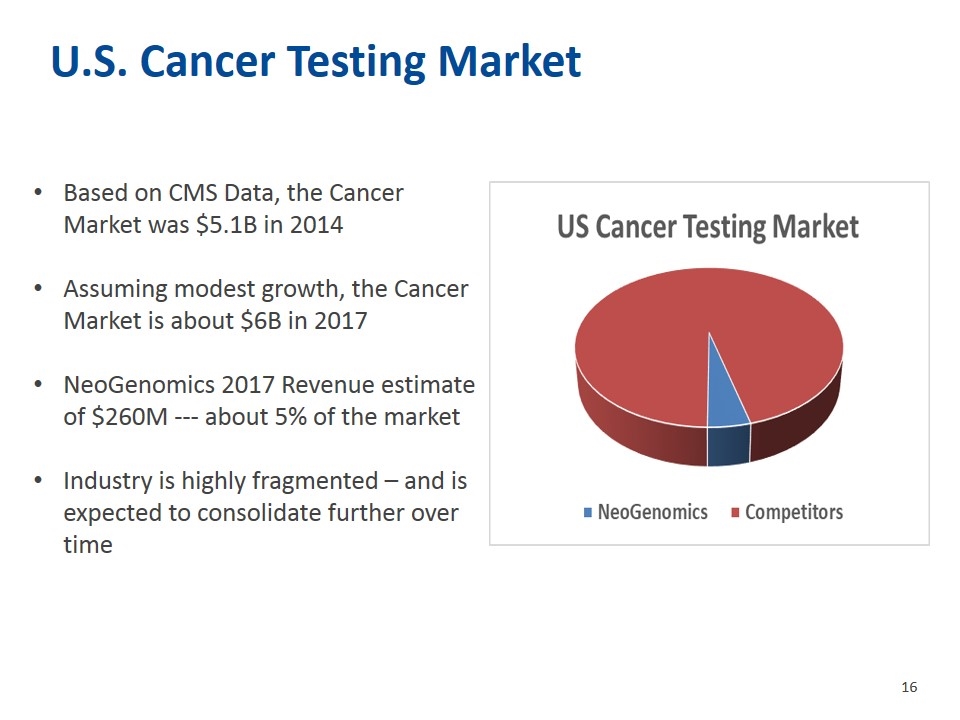
U.S. Cancer Testing Market Based on CMS Data, the Cancer Market was $5.1B in 2014 Assuming modest growth, the Cancer Market is about $6B in 2017 NeoGenomics 2017 Revenue estimate of $260M --- about 5% of the market Industry is highly fragmented – and is expected to consolidate further over time
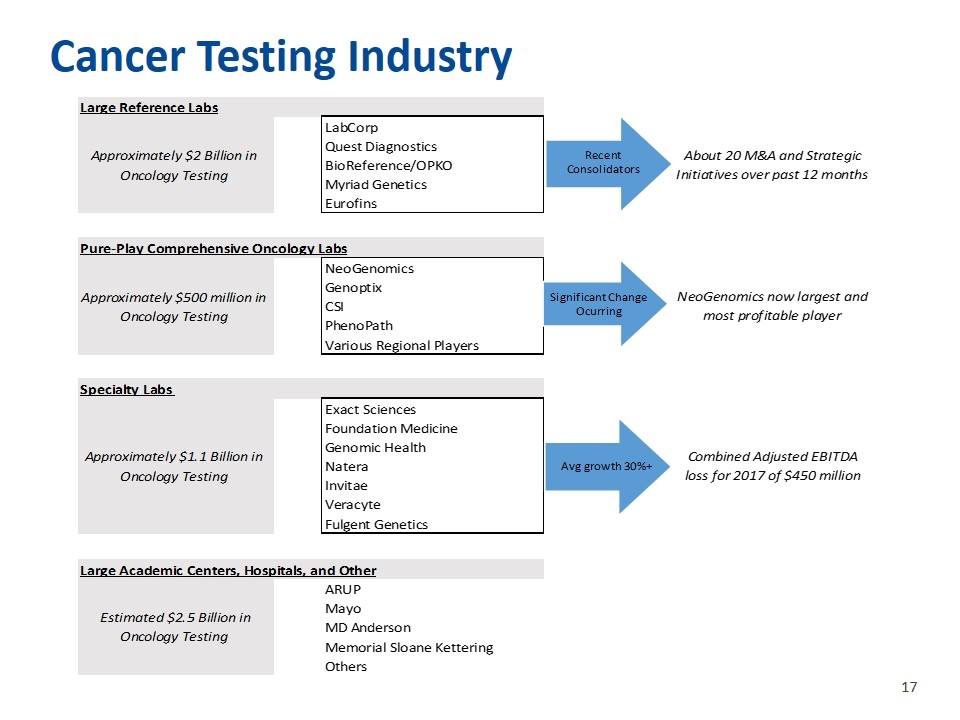
Cancer Testing Industry Large Reference Labs Approximately $2 Billion in Oncology Testing LabCorp Quest Diagnostics About 20 M&A and Strategic Initiatives over past 12 months BioReference/OPKO Myriad Genetics Eurofins Pure-Play Comprehensive Oncology Labs Approximately $500 million in Oncology Testing NeoGenomics Genoptix NeoGenomics now largest and most profitable player CSI PhenoPath Various Regional Players Specialty Labs Approximately $1.1 Billion in Oncology Testing Exact Sciences Foundation Medicine Genomic Health Combined Adjusted EBITDA loss for 2017 of $450 million Natera Invitae Veracyte Fulgent Genetics Large Academic Centers, Hospitals, and Other Estimated $2.5 Billion in Oncology Testing ARUP Mayo MD Anderson Memorial Sloane Kettering Others
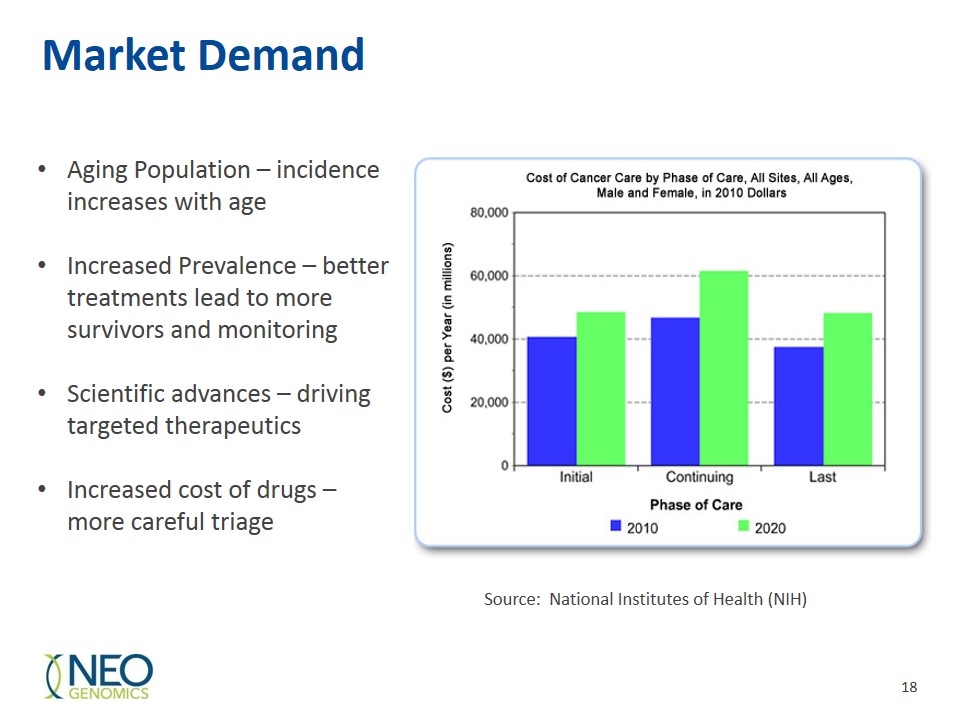
Market Demand Source: National Institutes of Health (NIH) Aging Population – incidence increases with age Increased Prevalence – better treatments lead to more survivors and monitoring Scientific advances – driving targeted therapeutics Increased cost of drugs – more careful triage
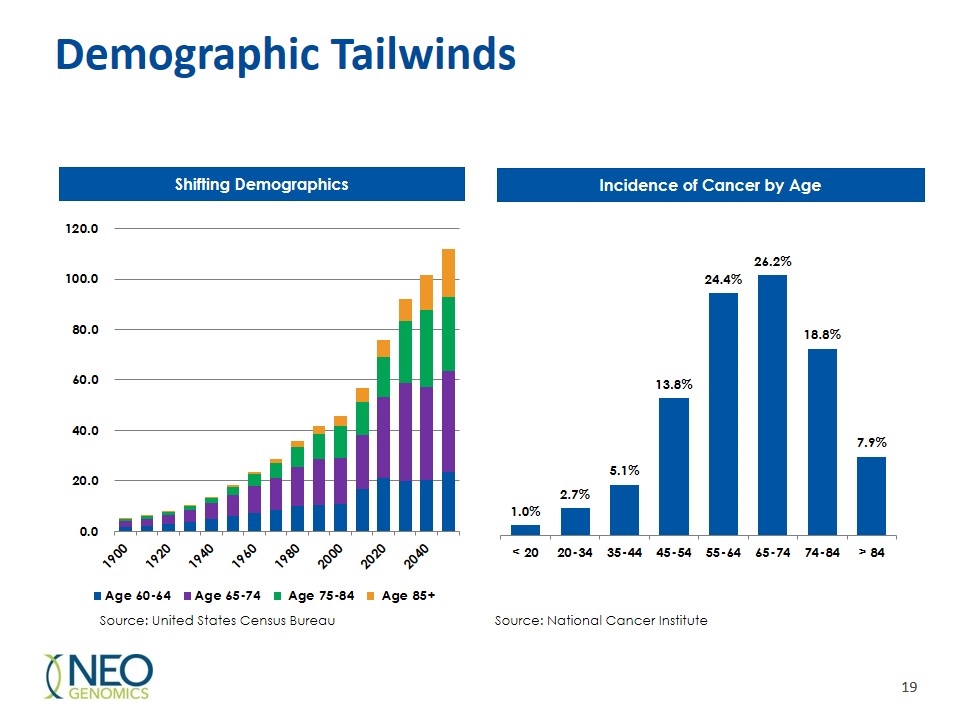
Demographic Tailwinds Shifting Demographics Incidence of Cancer by Age Source: United States Census Bureau Source: National Cancer Institute
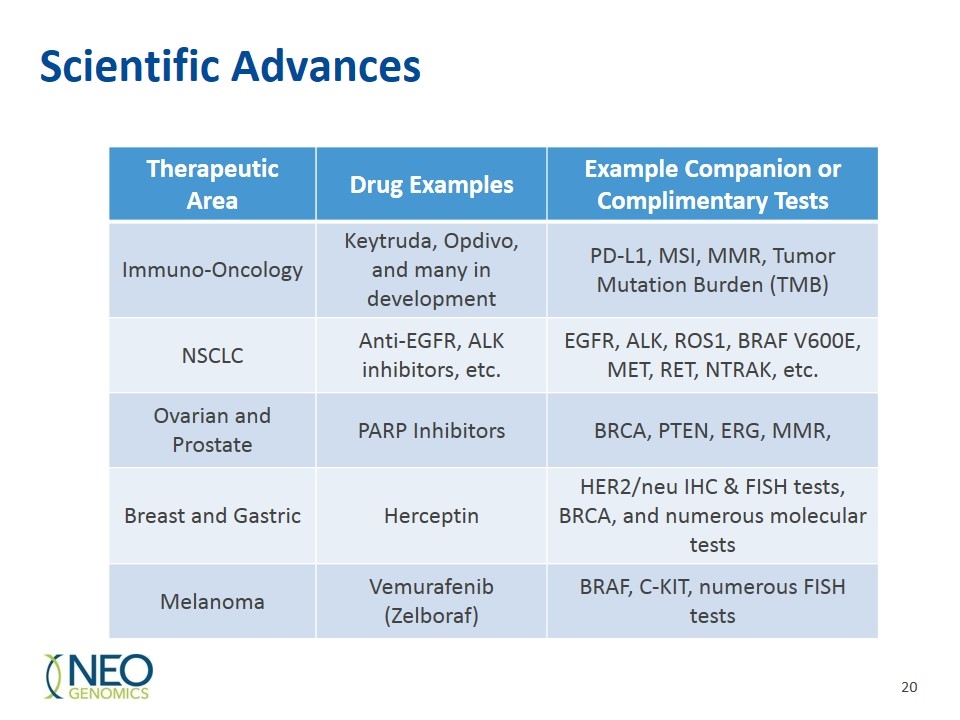
Scientific Advances Therapeutic Area Drug Examples Example Companion or Complimentary Tests Immuno-Oncology Keytruda, Opdivo, and many in development PD-L1, MSI, MMR, Tumor Mutation Burden (TMB) NSCLC Anti-EGFR, ALK inhibitors, etc. EGFR, ALK, ROS1, BRAF V600E, MET, RET, NTRAK, etc. Ovarian and Prostate PARP Inhibitors BRCA, PTEN, ERG, MMR, Breast and Gastric Herceptin HER2/neu IHC & FISH tests, BRCA, and numerous molecular tests Melanoma Vemurafenib (Zelboraf) BRAF, C-KIT, numerous FISH tests
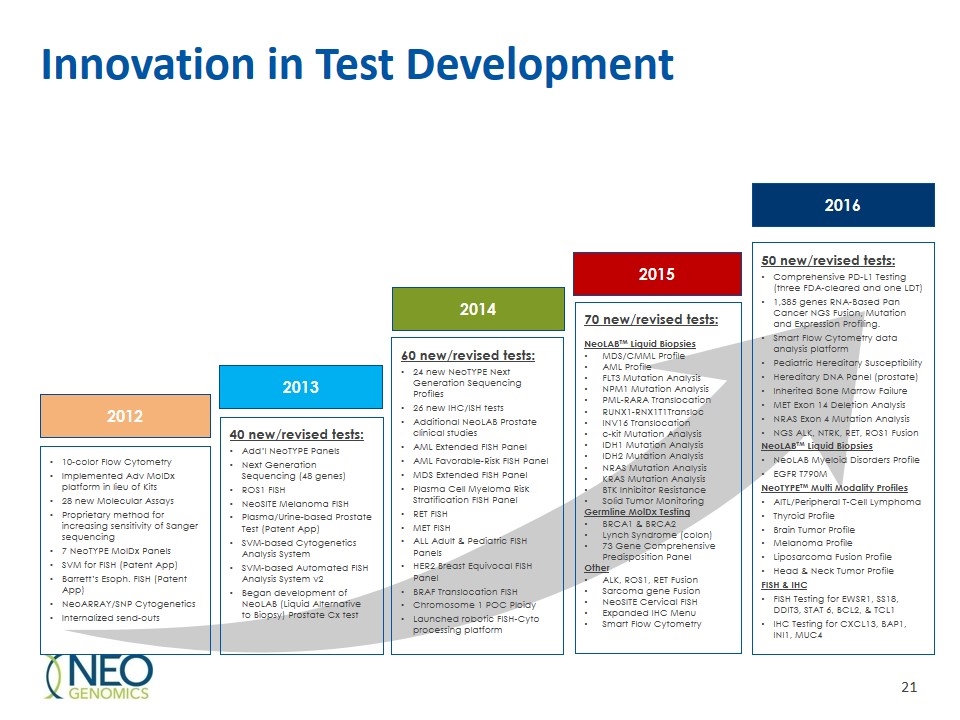
Innovation in Test Development 70 new/revised tests: NeoLABTM Liquid Biopsies MDS/CMML Profile AML Profile FLT3 Mutation Analysis NPM1 Mutation Analysis PML-RARA Translocation RUNX1-RNX1T1Transloc INV16 Translocation c-kit Mutation Analysis IDH1 Mutation Analysis IDH2 Mutation Analysis NRAS Mutation Analysis KRAS Mutation Analysis BTK Inhibitor Resistance Solid Tumor Monitoring Germline MolDx Testing BRCA1 & BRCA2 Lynch Syndrome (colon) 73 Gene Comprehensive Predisposition Panel Other ALK, ROS1, RET Fusion Sarcoma gene Fusion NeoSITE Cervical FISH Expanded IHC Menu Smart Flow Cytometry 10-color Flow Cytometry Implemented Adv MolDx platform in lieu of Kits 28 new Molecular Assays Proprietary method for increasing sensitivity of Sanger sequencing 7 NeoTYPE MolDx Panels SVM for FISH (Patent App) Barrett’s Esoph. FISH (Patent App) NeoARRAY/SNP Cytogenetics Internalized send-outs 40 new/revised tests: Add’l NeoTYPE Panels Next Generation Sequencing (48 genes) ROS1 FISH NeoSITE Melanoma FISH Plasma/Urine-based Prostate Test (Patent App) SVM-based Cytogenetics Analysis System SVM-based Automated FISH Analysis System v2 Began development of NeoLAB (Liquid Alternative to Biopsy) Prostate Cx test 60 new/revised tests: 24 new NeoTYPE Next Generation Sequencing Profiles 26 new IHC/ISH tests Additional NeoLAB Prostate clinical studies AML Extended FISH Panel AML Favorable-Risk FISH Panel MDS Extended FISH Panel Plasma Cell Myeloma Risk Stratification FISH Panel RET FISH MET FISH ALL Adult & Pediatric FISH Panels HER2 Breast Equivocal FISH Panel BRAF Translocation FISH Chromosome 1 POC Ploidy Launched robotic FISH-Cyto processing platform 50 new/revised tests: Comprehensive PD-L1 Testing (three FDA-cleared and one LDT) 1,385 genes RNA-Based Pan Cancer NGS Fusion, Mutation and Expression Profiling. Smart Flow Cytometry data analysis platform Pediatric Hereditary Susceptibility Hereditary DNA Panel (prostate) Inherited Bone Marrow Failure MET Exon 14 Deletion Analysis NRAS Exon 4 Mutation Analysis NGS ALK, NTRK, RET, ROS1 Fusion NeoLABTM Liquid Biopsies NeoLAB Myeloid Disorders Profile EGFR T790M NeoTYPETM Multi Modality Profiles AITL/Peripheral T-Cell Lymphoma Thyroid Profile Brain Tumor Profile Melanoma Profile Liposarcoma Fusion Profile Head & Neck Tumor Profile FISH & IHC FISH Testing for EWSR1, SS18, DDIT3, STAT 6, BCL2, & TCL1 IHC Testing for CXCL13, BAP1, INI1, MUC4 2012 2013 2014 2015 2016

Opportunities by Customer Type Pathologists & Hospitals (about 80% of Revenue) Large Market with 5,600 Hospitals in U.S., and 18,000 active Pathologists Enable Pathologists to practice using sophisticated tools/tests and “tech-only” services Unique ability to be “One-stop shop” with comprehensive oncology test menu Competitive pricing under contract, and agreements with hospitals & hospital GPOs Oncologists, Hematologists & Clinicians (about 10% of Revenue) Disease Panels, liquid biopsies, and comprehensive molecular menus Increasing opportunity to service larger practices with Partnership-based tech-only model Contracts with key Managed Care organizations Pharma Services & Other (about 10% of Revenue) Contract research/clinical trial support work for Pharma clients Opening in Geneva, Switzerland to handle European and Global studies MultiOmyx platform is a unique offering gaining acceptance by Pharma firms
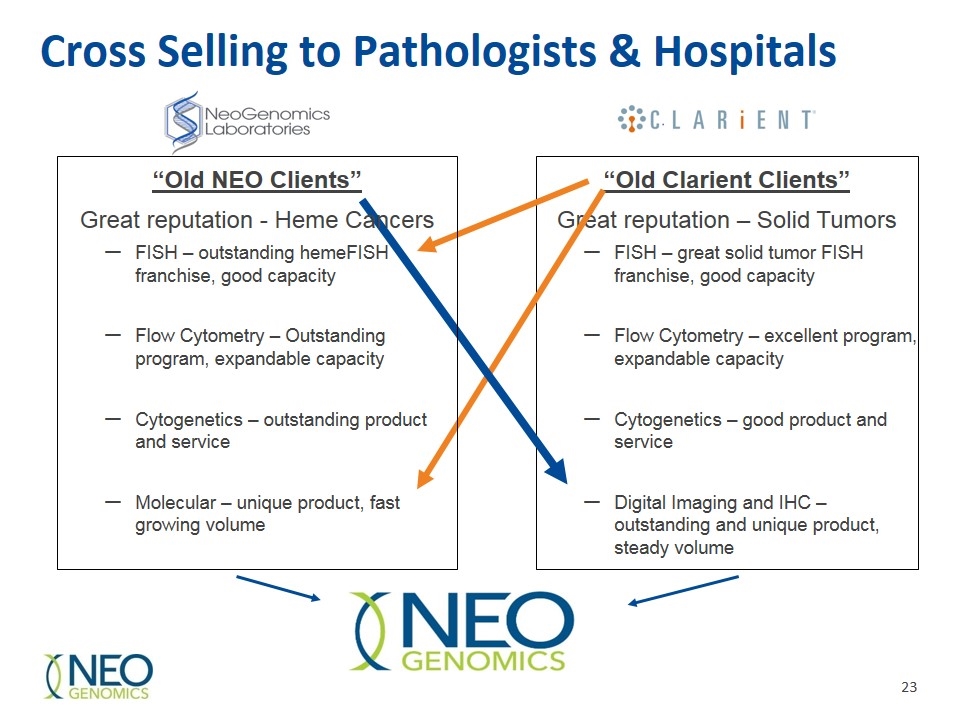
Cross Selling to Pathologists & Hospitals “Old Clarient Clients” Great reputation – Solid Tumors FISH – great solid tumor FISH franchise, good capacity Flow Cytometry – excellent program, expandable capacity Cytogenetics – good product and service Digital Imaging and IHC – outstanding and unique product, steady volume “Old NEO Clients” Great reputation - Heme Cancers FISH – outstanding hemeFISH franchise, good capacity Flow Cytometry – Outstanding program, expandable capacity Cytogenetics – outstanding product and service Molecular – unique product, fast growing volume

Cost Synergies and Opportunities 2015 Adjusted EBITDA NeoGenomics $ 9.7 MM Clarient Pro Forma $ 9.2 MM Pro Forma Combined $18.9 MM 2016 “New Neo” EBITDA$ 34.7 MM Clearly, “synergies” showed up in 2016 --- and there are more to be realized in 2017- 18 2017 – 2018 Opportunities: Laboratory facilities consolidated on 3/26/17 – teams now working as one Ability to run combined batches in So. California rather than two batches Ongoing supplier re-negotiations as contracts end – we have 2x volume Irvine laboratory closed on 4/30 – saving over $50k per month Cross selling now possible – accounts all one system Pharma services just starting to access Neo’s leading Molecular menu 9 + 9 equals 34
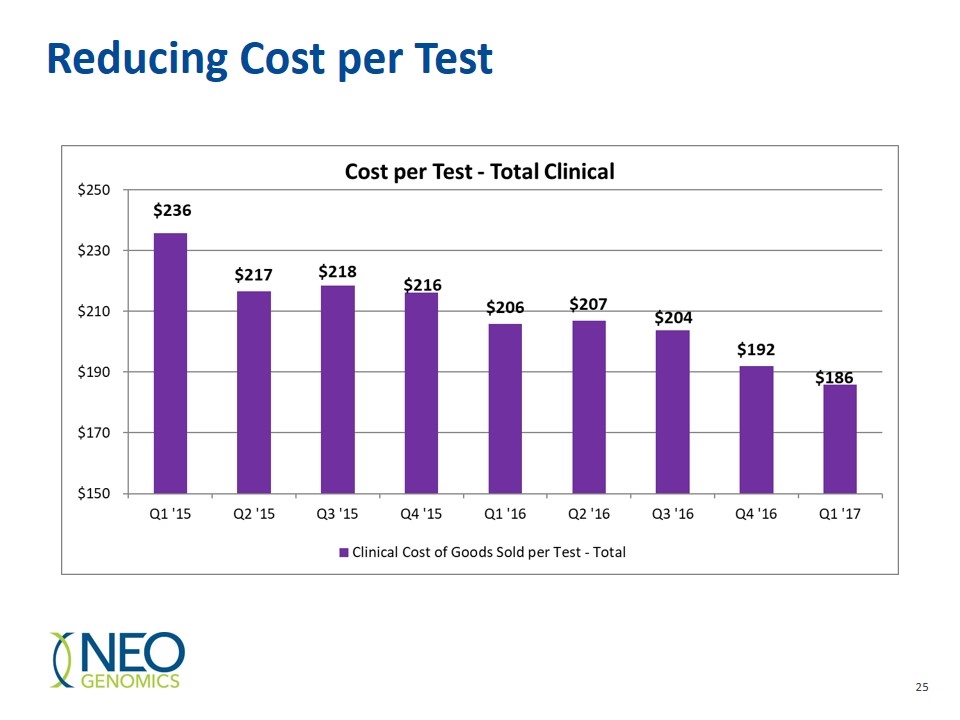
Reducing Cost per Test
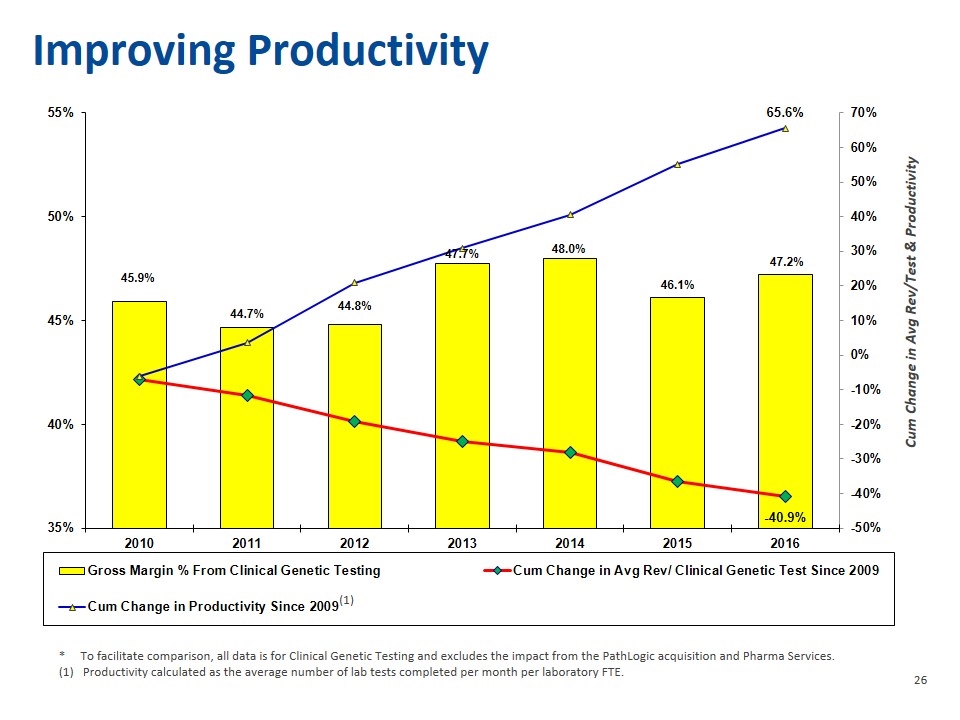
Improving Productivity * To facilitate comparison, all data is for Clinical Genetic Testing and excludes the impact from the PathLogic acquisition and Pharma Services. Productivity calculated as the average number of lab tests completed per month per laboratory FTE.
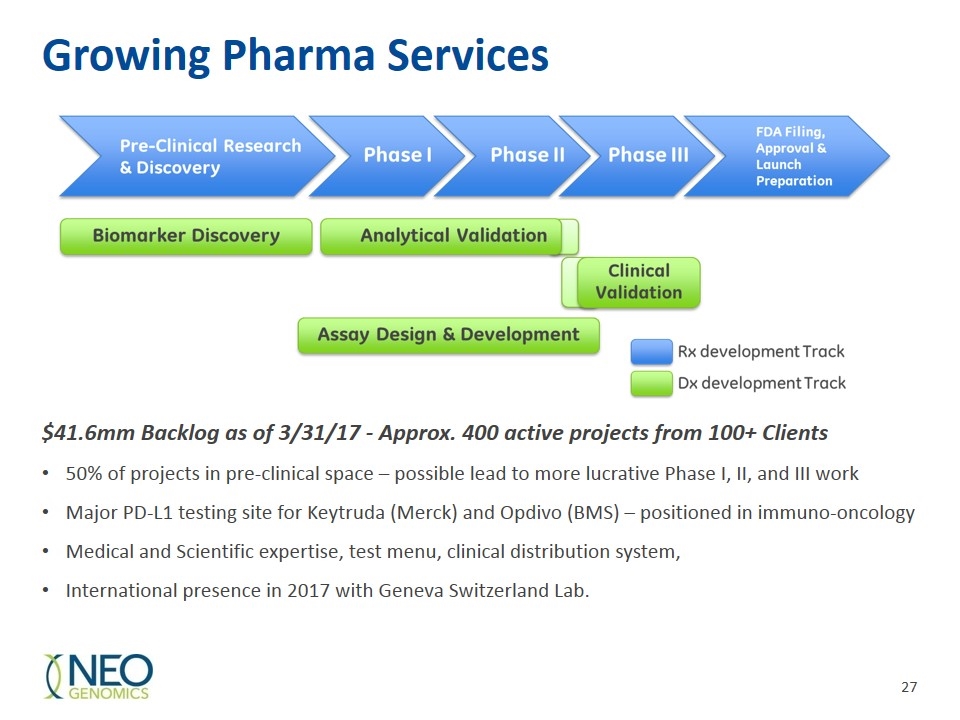
Growing Pharma Services $41.6mm Backlog as of 3/31/17 - Approx. 400 active projects from 100+ Clients 50% of projects in pre-clinical space – possible lead to more lucrative Phase I, II, and III work Major PD-L1 testing site for Keytruda (Merck) and Opdivo (BMS) – positioned in immuno-oncology Medical and Scientific expertise, test menu, clinical distribution system, International presence in 2017 with Geneva Switzerland Lab.
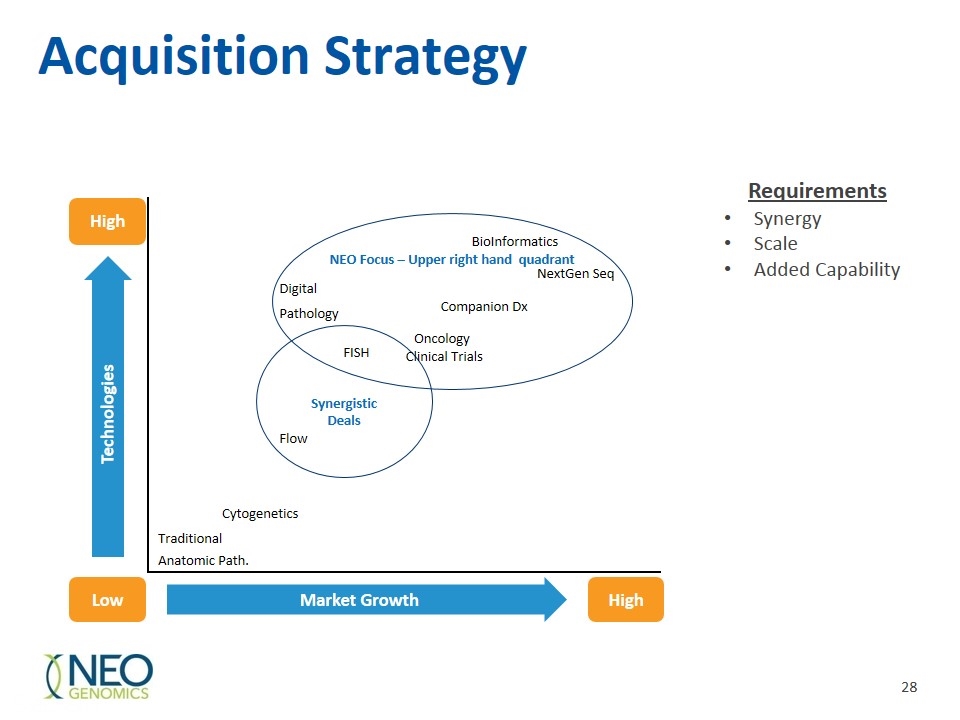
Acquisition Strategy NEO Focus – Upper right hand quadrant Synergistic Deals High High Low Market Growth Technologies NextGen Seq Digital Companion Dx Pathology Oncology FISH Clinical Trials Flow Cytogenetics Traditional Anatomic Path. BioInformatics Requirements Synergy Scale Added Capability
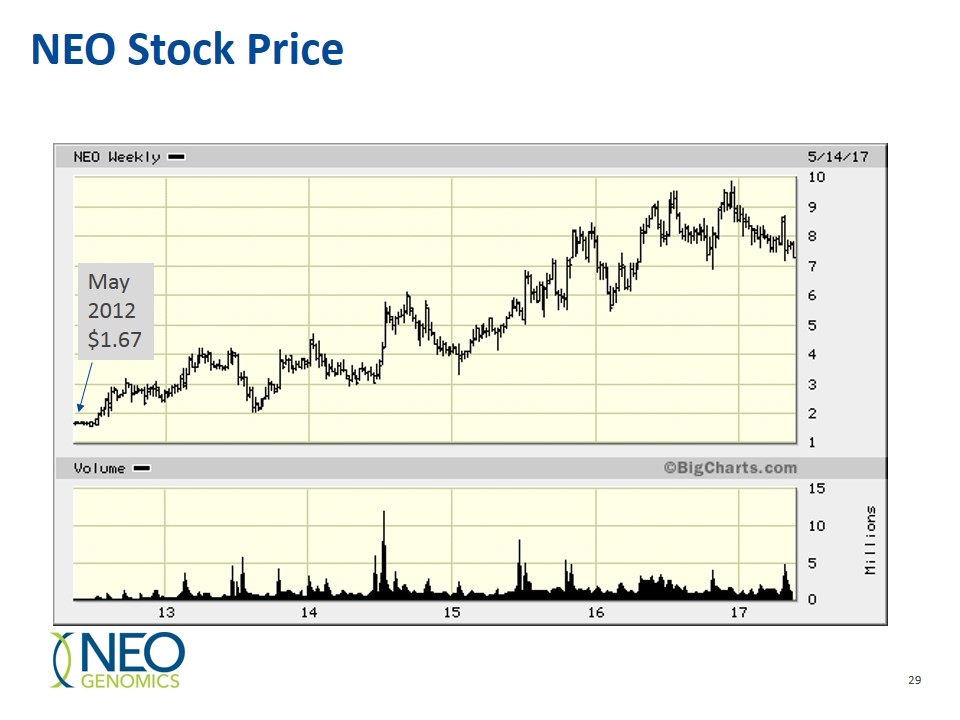
NEO Stock Price May 2012$1.67

2020 Goals By providing uncompromising quality, exceptional service & innovative solutions, we will be the world’s leading cancer testing and information company! 15%+ Clinical Volume CAGR 20%+ Pharma Services Revenue CAGR 20-25% Adjusted EBITDA Margins

Key Takeaways þUnique and leading pure-play Oncology Company þ History of strong performance þ Significant opportunities for growth þ Realization of cost synergies expected to accelerate þ Strategic opportunities highly likely over the next 36 mos þ Well positioned for continued Industry Leadership

Questions and Answers

Review of NEO Testing Platforms Lawrence M. Weiss, MD Medical Director, Aliso Viejo

Lawrence Weiss, M.D. Medical Director, Aliso Viejo Dr. Weiss currently serves as NeoGenomics’ Medical Director, Aliso Viejo and served as the Medical Director at Clarient Diagnostic Services, Inc. prior to the NeoGenomics acquisition in 2015. Dr. Weiss received his BS summa cum laude and MD summa cum laude from the University of Maryland. He completed a residency in Anatomic Pathology at the Brigham and Women’s Hospital in Boston, MA and a fellowship in surgical pathology at Stanford University Medical Center. He was previously an Assistant Professor at Stanford and Director of Surgical Pathology, President of the Medical Staff, and Chairman of Pathology at the City of Hope. He is the author of over 500 papers and book chapters, as well as over a dozen books, including an AFIP Lymph Node Fascicle, Applied Immunohistochemistry, Lymph Nodes, and the recently published Knowles’ Hematopathology. His laboratory discovered the first molecular evidence linking the Epstein-Barr virus with Hodgkin Lymphoma. He has won numerous awards, including the Benjamin Castleman, Arthur Purdy Stout, and the United States-Canadian Academy of Pathology Young Investigator Award, and has delivered over 250 national and international talks in pathology, including several named lectureships. He has been on the editorial board of ten scientific journals, and is a past President of the Los Angeles Society of Pathologists. He has been listed in the book The Best Doctors in America since 1994. Dr. Weiss’s diagnostic interests lie in lymph node pathology, adrenal pathology, tumor pathology, and immunohistochemistry.

Testing Modalities Offered by NeoGenomics Morphologic interpretation/consultation Immunohistochemistry Diagnostic Prognostic Flow cytometry Cytogenetics FISH Molecular studies Single gene Large NGS profiles
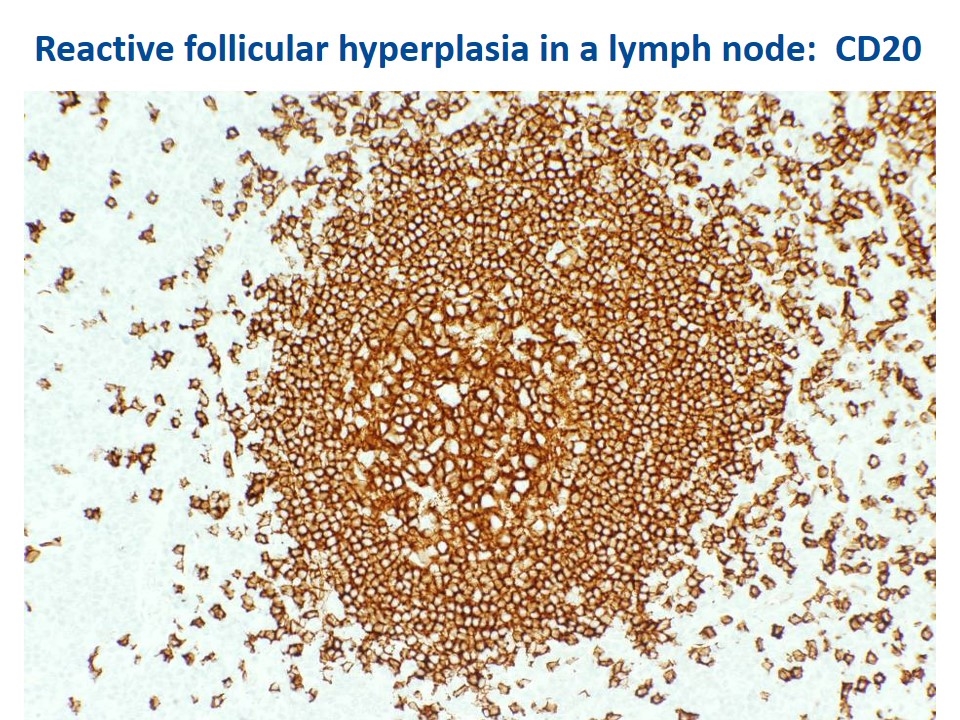
Reactive follicular hyperplasia in a lymph node: CD20
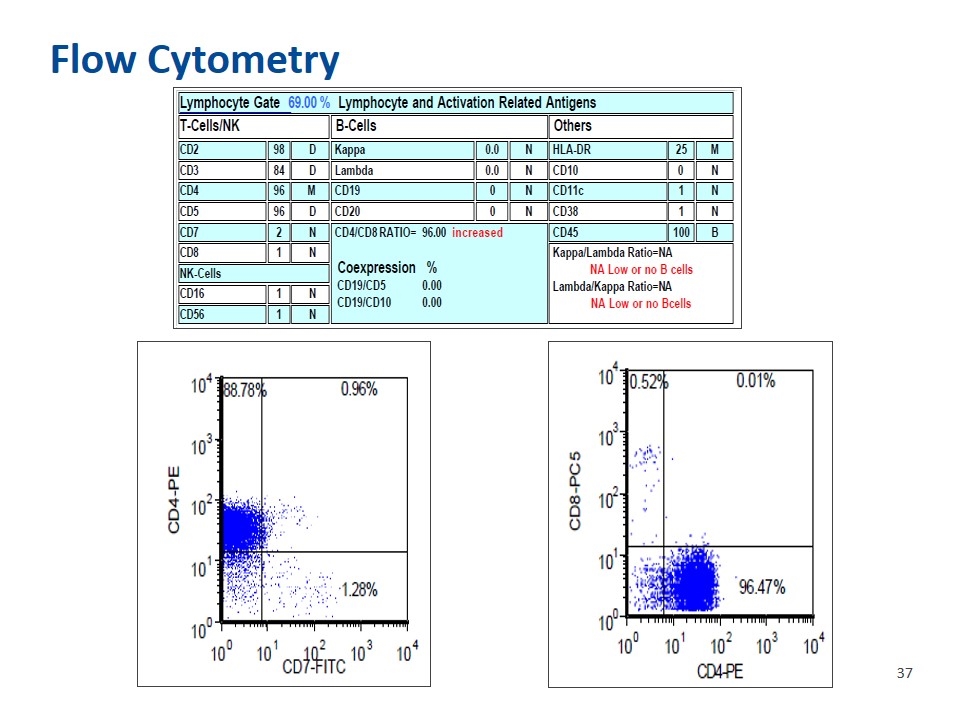
Flow Cytometry
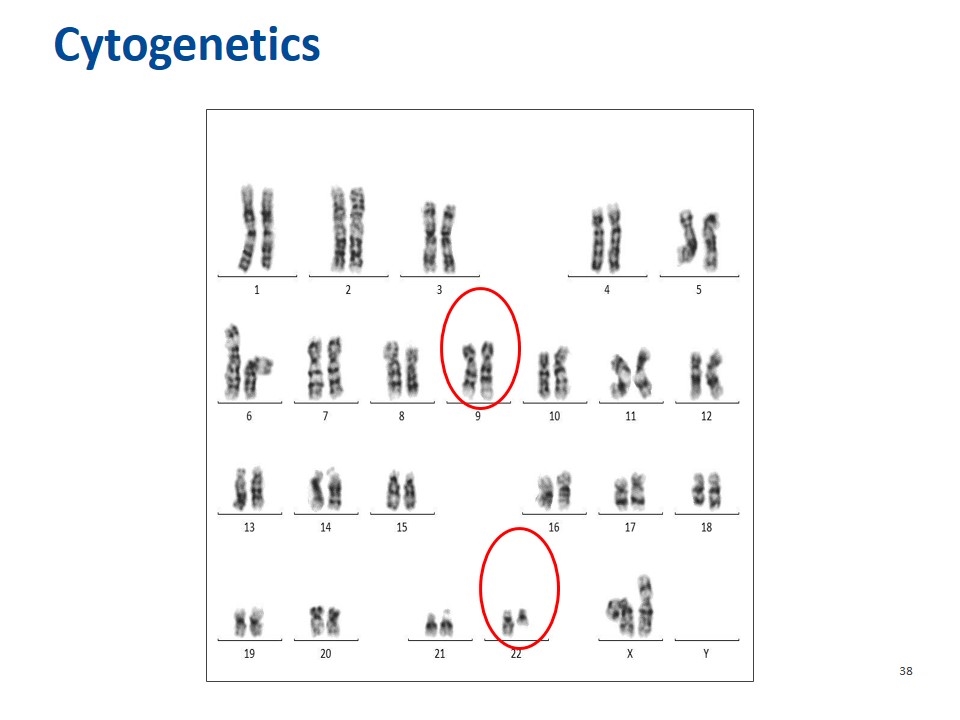
Cytogenetics
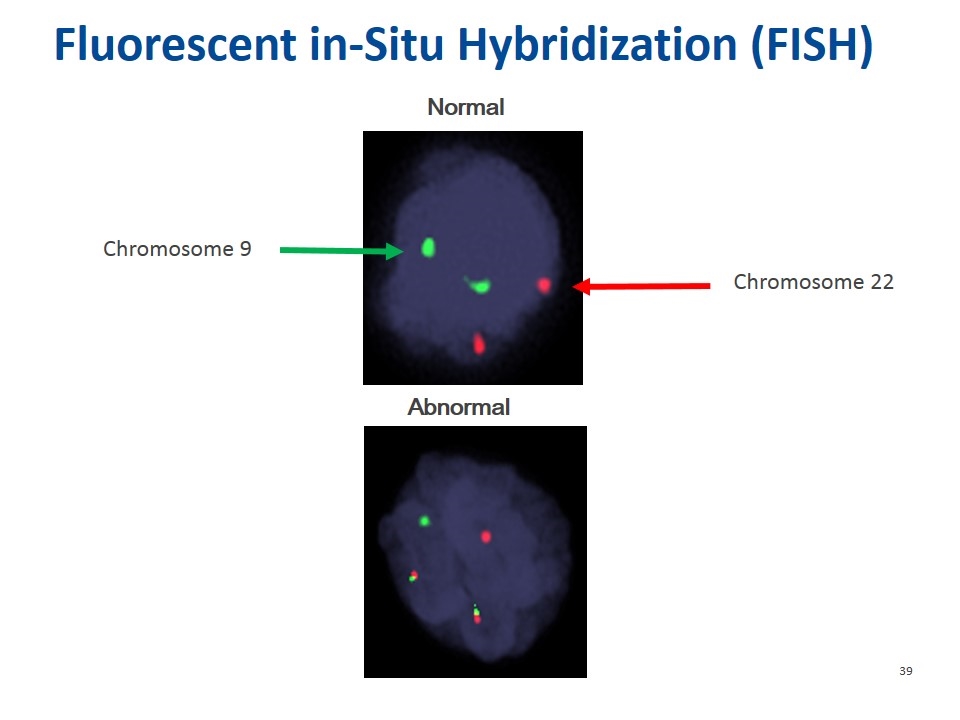
Normal Abnormal Fluorescent in-Situ Hybridization (FISH) Chromosome 9 Chromosome 22

Why Pathologists Choose NeoGenomics? Full service lab with most comprehensive menu. Highly experienced technical staff able to perform complex studies with high accuracy Flexible business models allows clients to perform professional interpretations Expert medical staff able to interpret any studies and answer any questions Superior turn-around times Culture of compliance

PD-L1 Testing Lawrence M. Weiss, MD Medical Director, Aliso Viejo

Dual Role of Immune System in Cancer Suppress neoplastic growth by recognizing cells with neoantigens and eliminating them Promote neoplastic growth by inadvertently selecting for cancer clones that evade immune surveillance
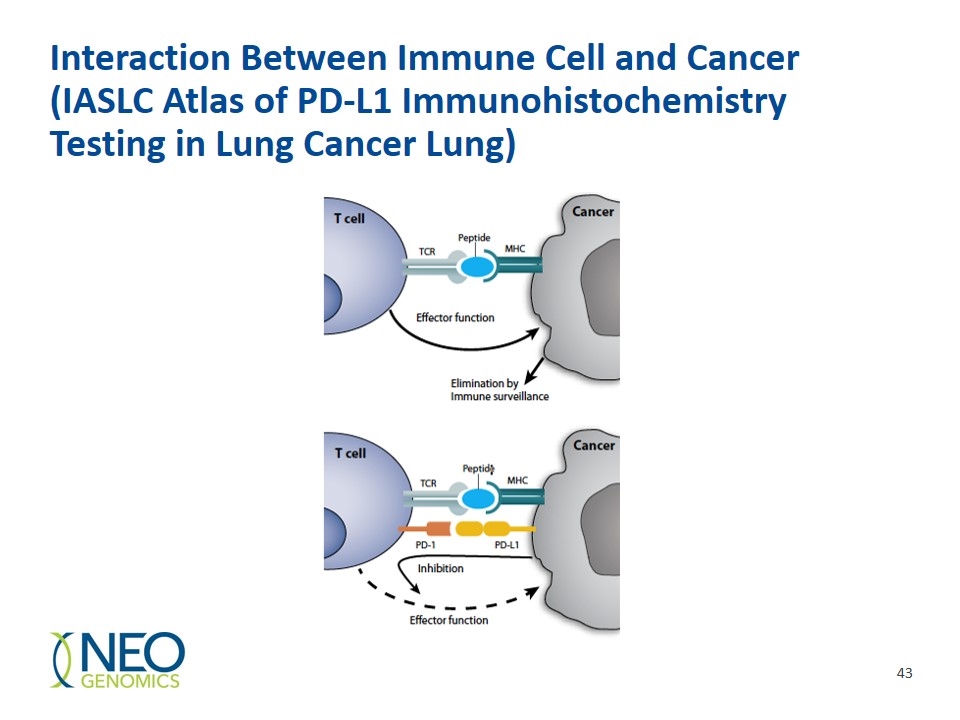
Interaction Between Immune Cell and Cancer (IASLC Atlas of PD-L1 Immunohistochemistry Testing in Lung Cancer Lung)
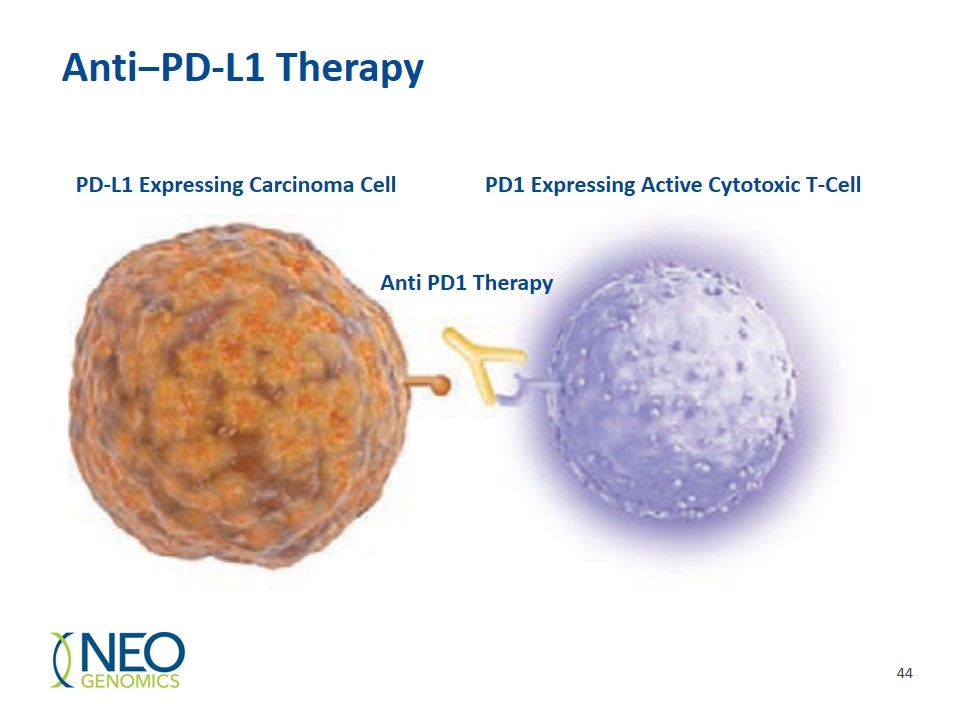
Anti‒PD-L1 Therapy PD-L1 Expressing Carcinoma Cell PD1 Expressing Active Cytotoxic T-Cell Anti PD1 Therapy
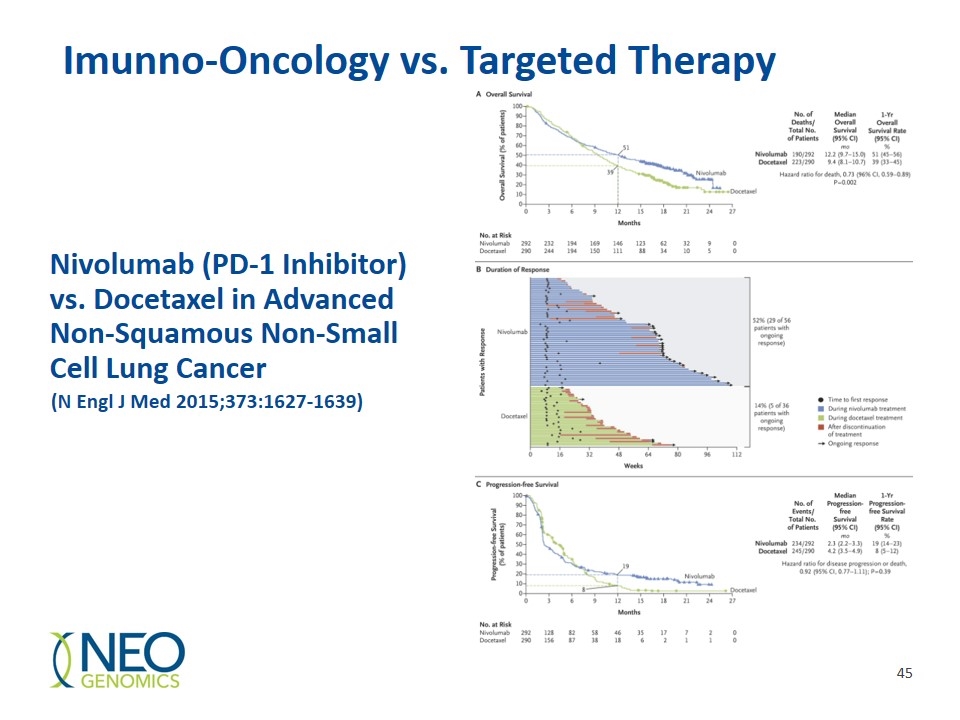
Nivolumab (PD-1 Inhibitor) vs. Docetaxel in Advanced Non-Squamous Non-Small Cell Lung Cancer (N Engl J Med 2015;373:1627-1639) Imunno-Oncology vs. Targeted Therapy
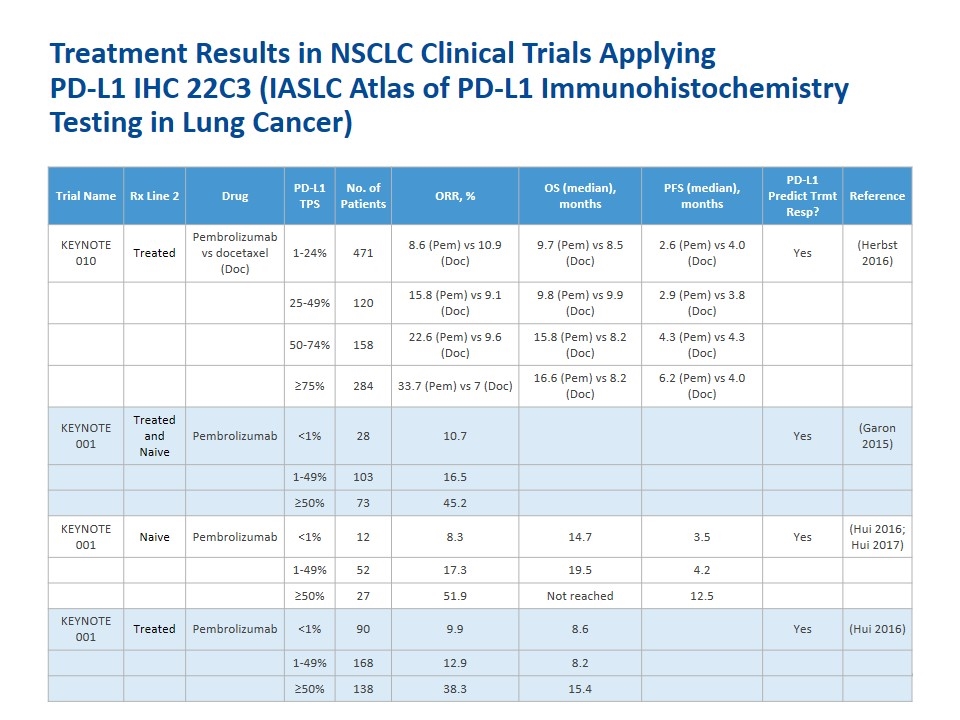
Treatment Results in NSCLC Clinical Trials Applying PD-L1 IHC 22C3 (IASLC Atlas of PD-L1 Immunohistochemistry Testing in Lung Cancer) Trial Name Rx Line 2 Drug PD-L1 TPS No. of Patients ORR, % OS (median), months PFS (median), months PD-L1 Predict Trmt Resp? Reference KEYNOTE 010 Treated Pembrolizumab vs docetaxel (Doc) 1-24% 471 8.6 (Pem) vs 10.9 (Doc) 9.7 (Pem) vs 8.5 (Doc) 2.6 (Pem) vs 4.0 (Doc) Yes (Herbst 2016) 25-49% 120 15.8 (Pem) vs 9.1 (Doc) 9.8 (Pem) vs 9.9 (Doc) 2.9 (Pem) vs 3.8 (Doc) 50-74% 158 22.6 (Pem) vs 9.6 (Doc) 15.8 (Pem) vs 8.2 (Doc) 4.3 (Pem) vs 4.3 (Doc) ≥75% 284 33.7 (Pem) vs 7 (Doc) 16.6 (Pem) vs 8.2 (Doc) 6.2 (Pem) vs 4.0 (Doc) KEYNOTE 001 Treated and Naive Pembrolizumab <1% 28 10.7 Yes (Garon 2015) 1-49% 103 16.5 ≥50% 73 45.2 KEYNOTE 001 Naive Pembrolizumab <1% 12 8.3 14.7 3.5 Yes (Hui 2016; Hui 2017) 1-49% 52 17.3 19.5 4.2 ≥50% 27 51.9 Not reached 12.5 KEYNOTE 001 Treated Pembrolizumab <1% 90 9.9 8.6 Yes (Hui 2016) 1-49% 168 12.9 8.2 ≥50% 138 38.3 15.4
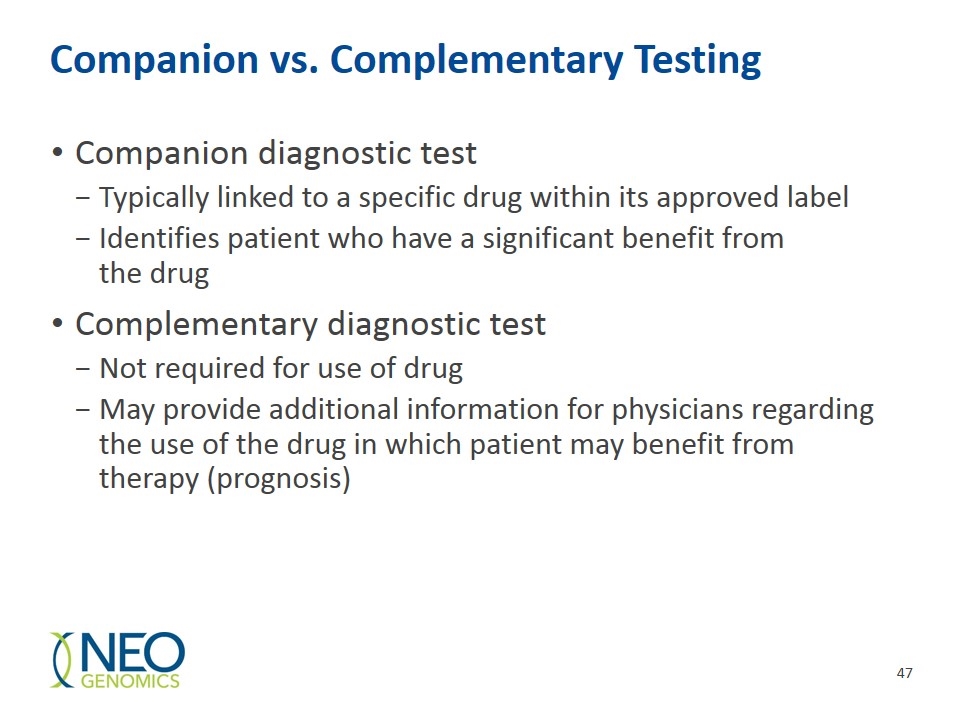
Companion vs. Complementary Testing Companion diagnostic test Typically linked to a specific drug within its approved label Identifies patient who have a significant benefit from the drug Complementary diagnostic test Not required for use of drug May provide additional information for physicians regarding the use of the drug in which patient may benefit from therapy (prognosis)

PD-L1 Testing Companies develop an IHC kit to be used as a companion or complementary diagnostic test for each drug Different kits, with different antibodies and different scoring systems Different manufacturers (DAKO vs. Ventana) Different machine platforms Count tumor cells, inflammatory cells, or both
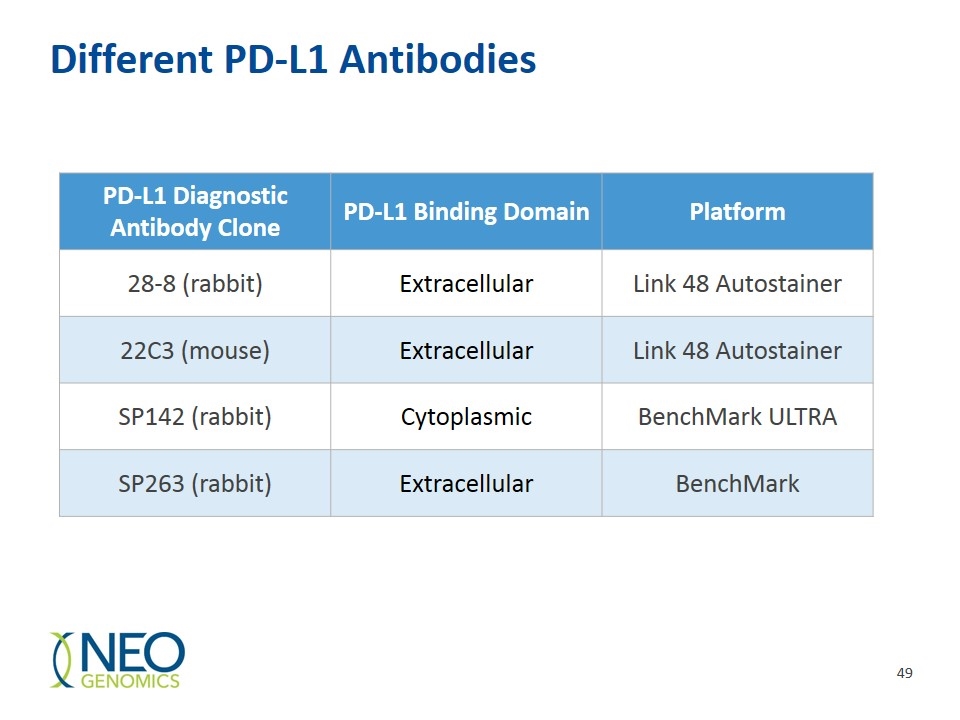
Different PD-L1 Antibodies PD-L1 Diagnostic Antibody Clone PD-L1 Binding Domain Platform 28-8 (rabbit) Extracellular Link 48 Autostainer 22C3 (mouse) Extracellular Link 48 Autostainer SP142 (rabbit) Cytoplasmic BenchMark ULTRA SP263 (rabbit) Extracellular BenchMark
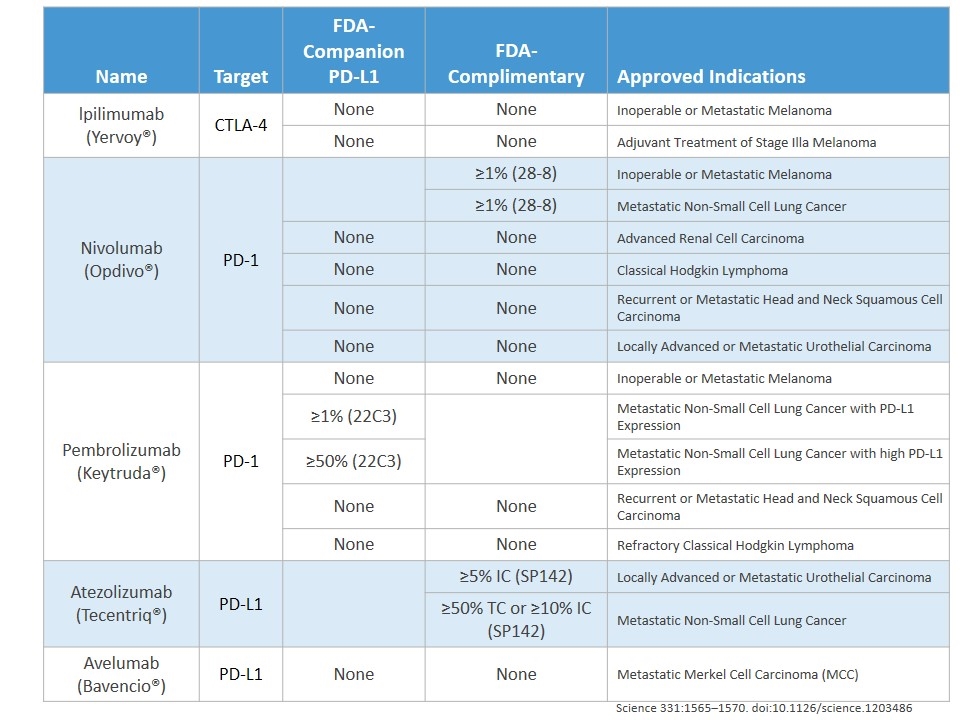
PD-1/PD-L1/CTLA-4 Treatment Name Target FDA-Companion PD-L1 FDA-Complimentary Approved Indications lpilimumab (Yervoy®) CTLA-4 None None Inoperable or Metastatic Melanoma None None Adjuvant Treatment of Stage Illa Melanoma Nivolumab (Opdivo®) PD-1 ≥1% (28-8) Inoperable or Metastatic Melanoma ≥1% (28-8) Metastatic Non-Small Cell Lung Cancer None None Advanced Renal Cell Carcinoma None None Classical Hodgkin Lymphoma None None Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma None None Locally Advanced or Metastatic Urothelial Carcinoma Pembrolizumab (Keytruda®) PD-1 None None Inoperable or Metastatic Melanoma ≥1% (22C3) Metastatic Non-Small Cell Lung Cancer with PD-L1 Expression ≥50% (22C3) Metastatic Non-Small Cell Lung Cancer with high PD-L1 Expression None None Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma None None Refractory Classical Hodgkin Lymphoma Atezolizumab (Tecentriq®) PD-L1 ≥5% IC (SP142) Locally Advanced or Metastatic Urothelial Carcinoma ≥50% TC or ≥10% IC (SP142) Metastatic Non-Small Cell Lung Cancer Avelumab (Bavencio®) PD-L1 None None Metastatic Merkel Cell Carcinoma (MCC) Science 331:1565–1570. doi:10.1126/science.1203486
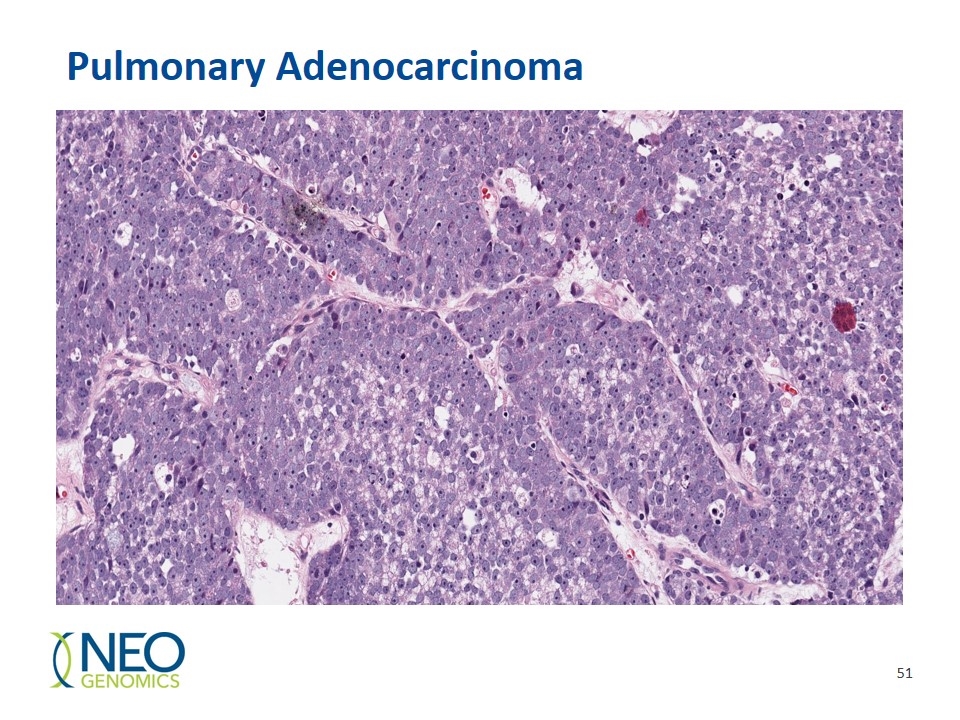
Pulmonary Adenocarcinoma
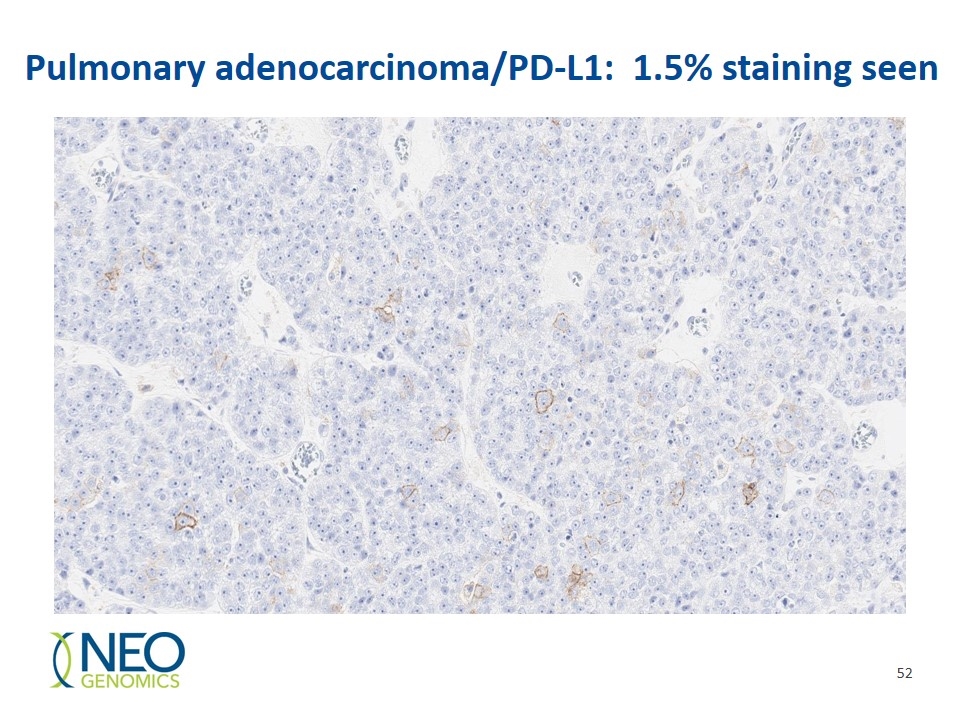
Pulmonary adenocarcinoma/PD-L1: 1.5% staining seen
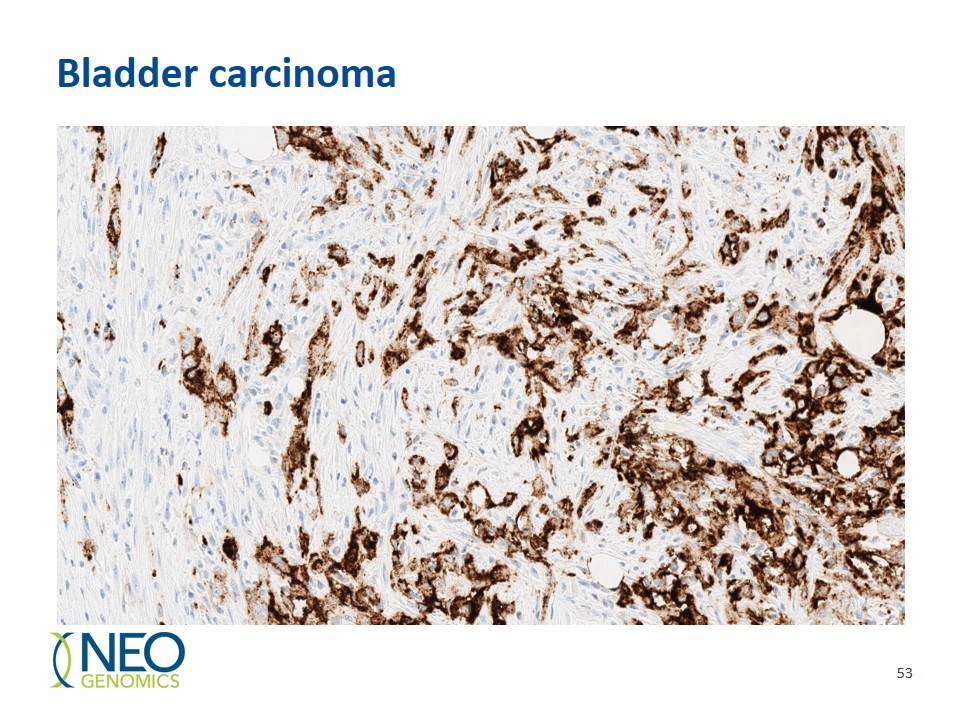
Bladder carcinoma
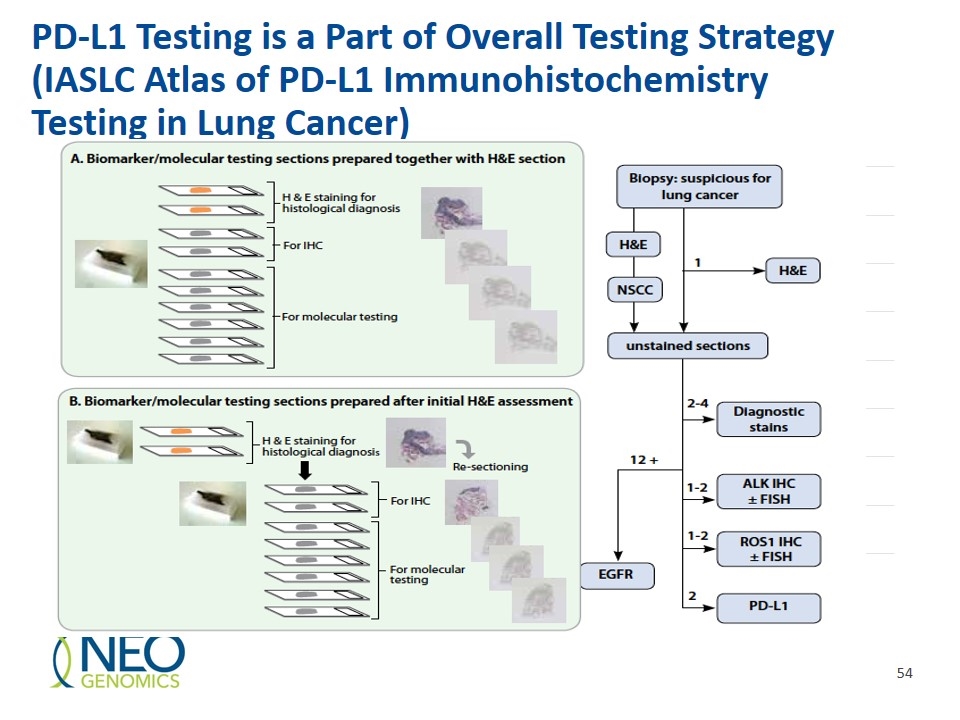
PD-L1 Testing is a Part of Overall Testing Strategy (IASLC Atlas of PD-L1 Immunohistochemistry Testing in Lung Cancer)
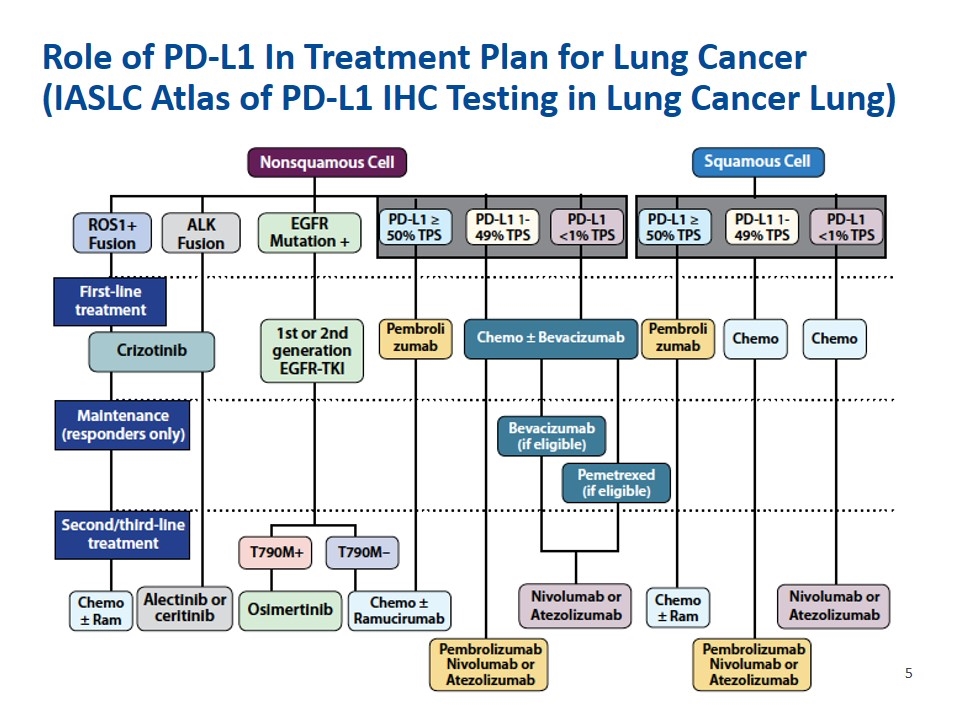
Role of PD-L1 In Treatment Plan for Lung Cancer (IASLC Atlas of PD-L1 IHC Testing in Lung Cancer Lung)

PD-1 Therapy Companion Testing: Rapidly Changing Landscape Keytruda approved on May 23, 2017 for use in unresectable or metastatic solid tumors that have been identified as having microsatellite instability-high (MSI-H) or mismatch repair deficient (dMMR) We already do routine testing for MSI/MMR in colon and endometrial carcinoma This new indication will open up MSI/MMR testing for other major cancers, including other gastrointestinal tumors , breast, prostate, bladder, thyroid, etc.

Key Takeaways þ Full-service Lab with complete range of testing in all major testing modalities - “one-stop” shop þ High level of technical and professional expertise in all cancer testing areas þ PD-L1 and related testing is an important new area and will continue to grow in the foreseeable future þ Well positioned to address rapidly evolving cancer landscape

Questions and Answers

15 Minute Break The Webcast will resume at approximately 10:15 AM PST

Pharma Services Overview Gina Wallar, PhD, MPH Division Vice President, Pharma Services Sales

Gina Wallar, PhD, MPH Division Vice President, Pharma Services Sales Gina Wallar currently serves as Division Vice President for Pharma Services Sales at NeoGenomics, a leading provider of specialty testing services and solutions for clinical trials. NeoGenomics Pharma Services provides customized biomarker testing services to meet sponsors and researchers’ requirements and leverages technologies such as immunohistochemistry, florescence in situ hybridization, flow cytometry, and a wide array of molecular services. Previously, she served as the Director for LabCorp Clinical Trials (formerly Genzyme Genetics) in Los Angeles, CA for over 12 years and oversaw PD-L1 testing supporting the first companion diagnostic approval. The CLIA/CAP laboratory focused in custom testing with an emphasis on companion diagnostics solutions in oncology. Gina earned her PhD at UCLA in cancer epidemiology with her field of study on risk susceptibility polymorphisms in the cancer stem cell pathway. Her research interests include risk factors for identifying susceptibility to cancer as well as molecular prognostic indicators of disease state.

Highlights Large and rapidly growing market opportunity Early successes and gaining momentum Immuno-oncology is a major driver of growth potential Investing for growth Unique and proprietary capabilities Expanding internationally
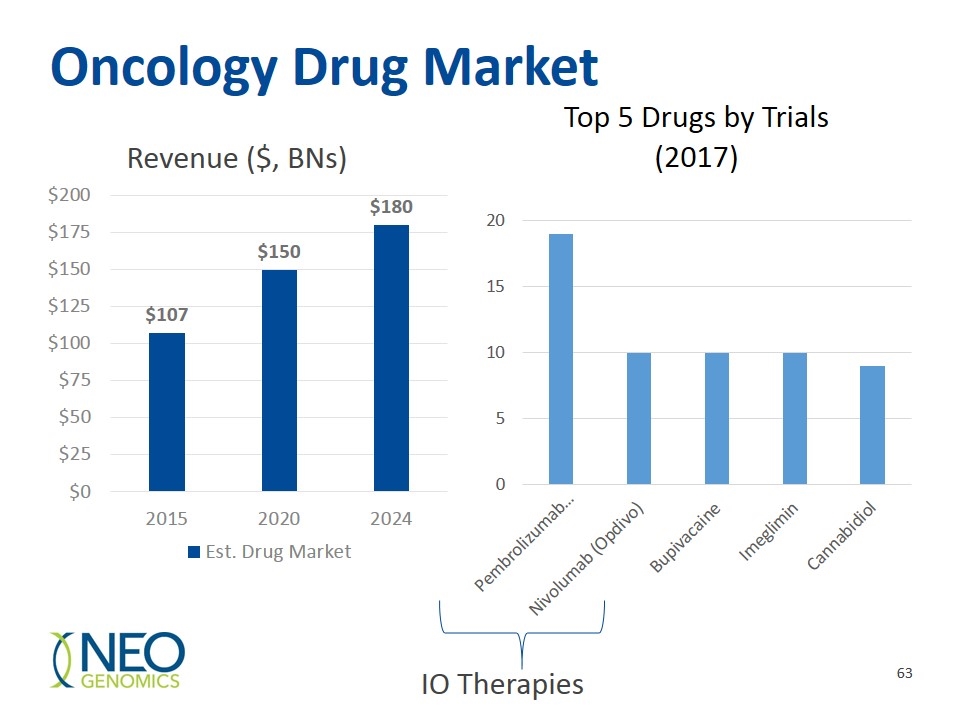
Oncology Drug Market IO Therapies

Clinical Trial Landscape In 2016: 14,239 trials began world wide (27% oncology) 5,223 trials began by US companies (33% oncology) 4,047 trials began by EU companies (24% oncology) Global market size for central lab testing services for trials: $2.4 - $3.5 BN LEK Analysis 2014 226 # of U.S. Trials: Ph 1 Ph 2 Ph 3 977 1,826
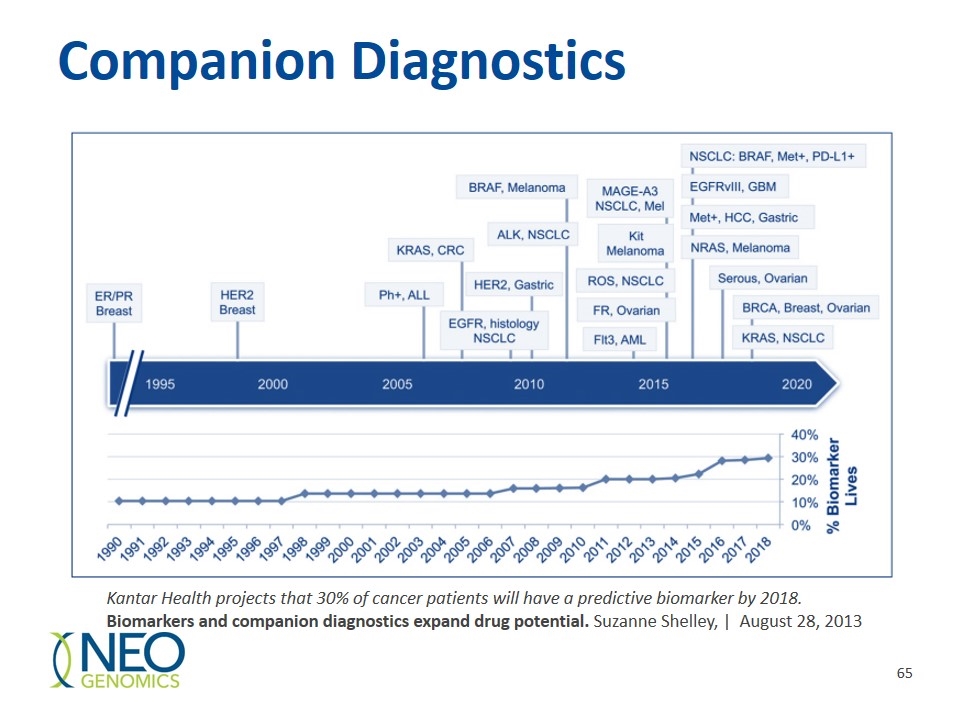
Companion Diagnostics Kantar Health projects that 30% of cancer patients will have a predictive biomarker by 2018. Biomarkers and companion diagnostics expand drug potential. Suzanne Shelley, | August 28, 2013

Neo Pharma Services Growth Drivers Companion and Complementary Diagnostics Immuno-Oncology MultiOmyx Molecular High Quality Sales Personnel International expansion
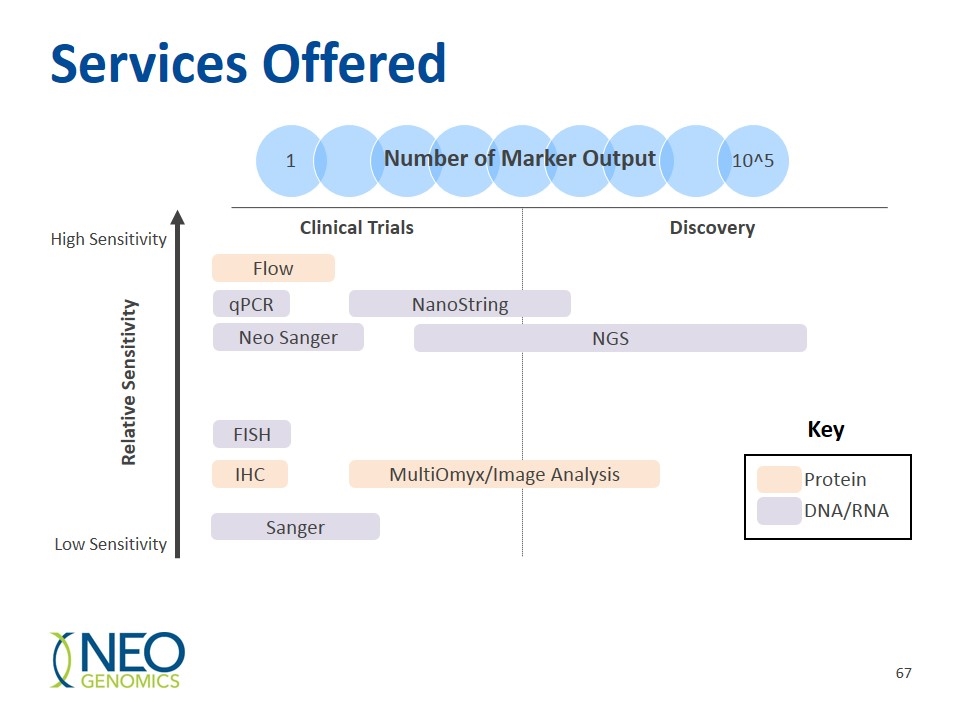
Services Offered Number of Marker Output qPCR Flow IHC FISH NGS Relative Sensitivity Discovery MultiOmyx/Image Analysis Neo Sanger Sanger Clinical Trials Protein DNA/RNA Key High Sensitivity Low Sensitivity NanoString 1 10^5
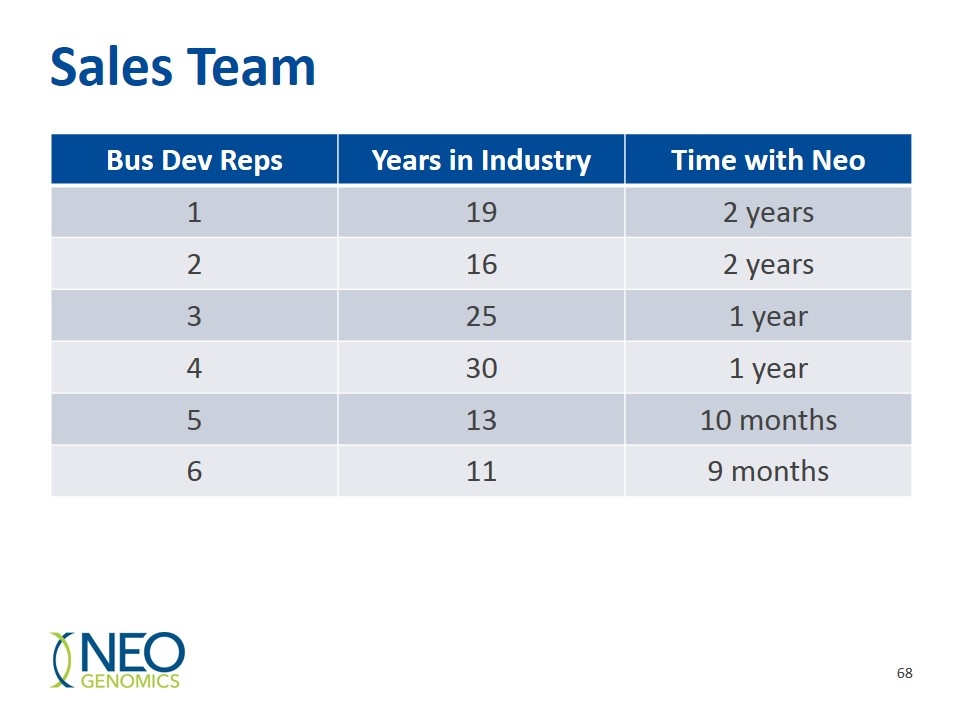
Sales Team Bus Dev Reps Years in Industry Time with Neo 1 19 2 years 2 16 2 years 3 25 1 year 4 30 1 year 5 13 10 months 6 11 9 months
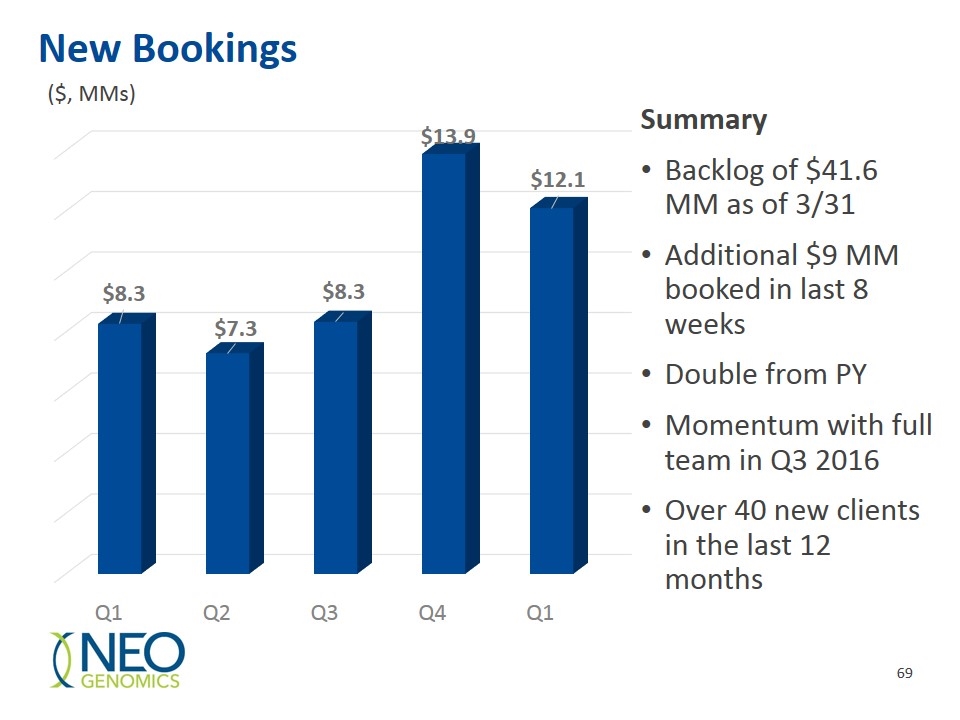
New Bookings Summary Backlog of $41.6 MM as of 3/31 Additional $9 MM booked in last 8 weeks Double from PY Momentum with full team in Q3 2016 Over 40 new clients in the last 12 months ($, MMs)
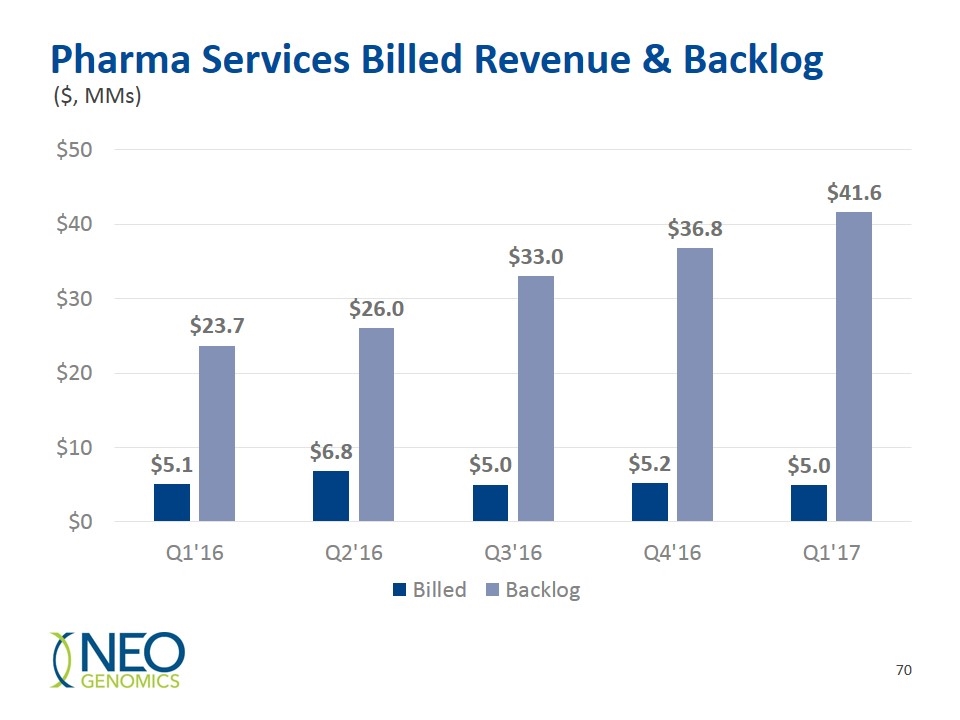
Pharma Services Billed Revenue & Backlog ($, MMs)

Recent Companion Diagnostic Success Selected by Merck due to IHC expertise Participated in Early Validation Program for Keytruda One of only three labs to offer PD-L1 testing on Day 1 Now, leading provider of clinical PD-L1 testing
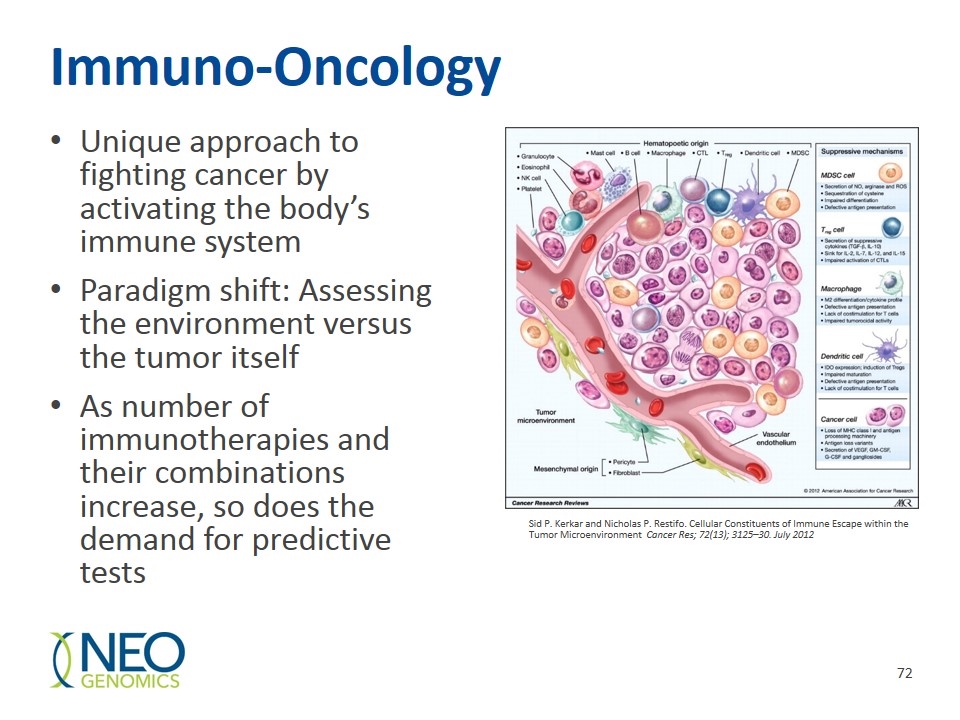
Immuno-Oncology Unique approach to fighting cancer by activating the body’s immune system Paradigm shift: Assessing the environment versus the tumor itself As number of immunotherapies and their combinations increase, so does the demand for predictive tests Sid P. Kerkar and Nicholas P. Restifo. Cellular Constituents of Immune Escape within the Tumor Microenvironment Cancer Res; 72(13); 3125–30. July 2012
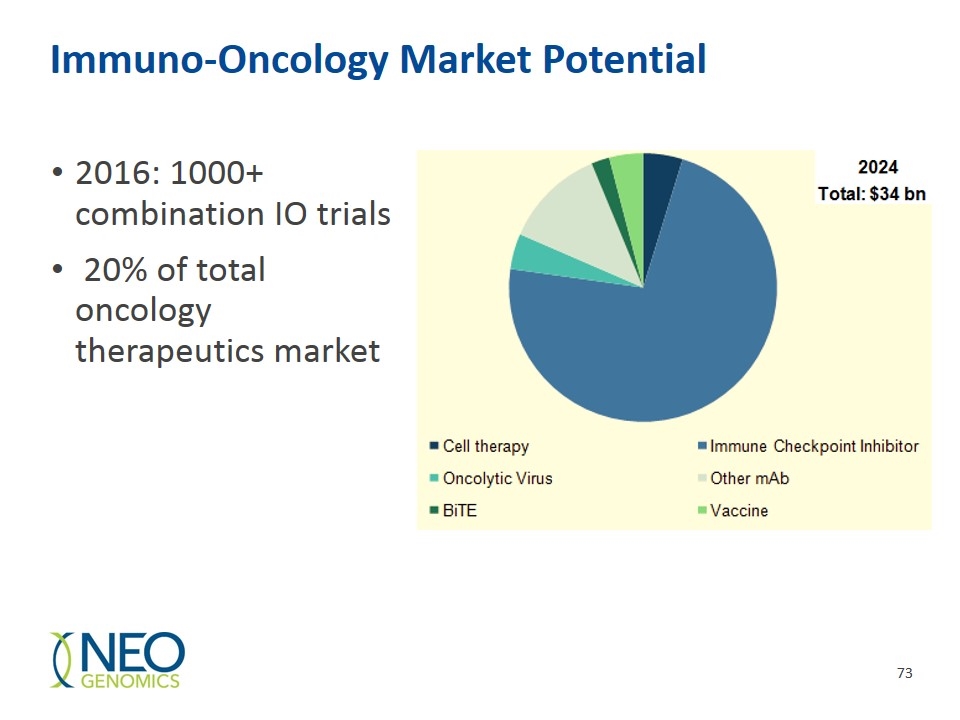
Immuno-Oncology Market Potential 2016: 1000+ combination IO trials 20% of total oncology therapeutics market

Pharma Activity in Immuno-Oncology Company Name IO Therapy IO Pipeline 2016 Oncology Sales ($, MMs) F. Hoffmann-La Roche Ltd Tecentriq >20 compounds $27,106 Celgene Corp > 8 Programs 10,011 Novartis AG 6 compounds 8,651 AstraZeneca Plc Imfinzi 27 trials 3,279 Bristol-Myers Squibb Opdivo; Yervoy > 10 compounds 6,801 Pfizer Inc Bavencio 16 trials 4,458 AbbVie Inc 4 compounds 2,653 Merck & Co Inc Keytruda 34 trials 1,685 Johnson & Johnson Darzalex 3 programs 5,307 Amgen Inc Imlygic 8 programs 2,947 Source: GlobalData
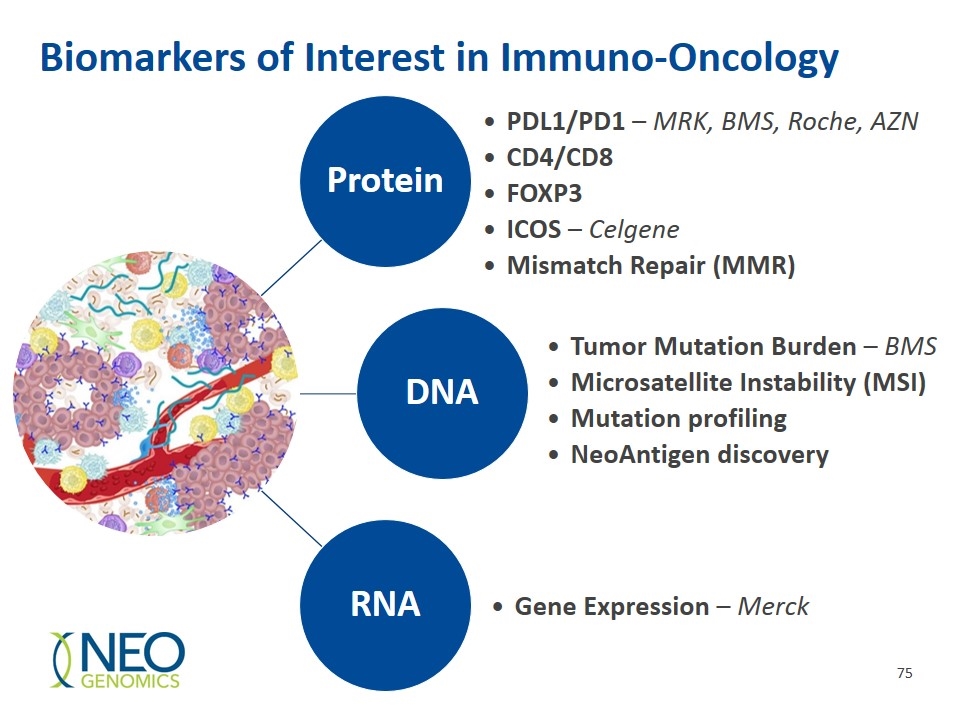
Biomarkers of Interest in Immuno-Oncology PDL1/PD1 – MRK, BMS, Roche, AZN CD4/CD8 FOXP3 ICOS – Celgene Mismatch Repair (MMR) Tumor Mutation Burden – BMS Microsatellite Instability (MSI) Mutation profiling NeoAntigen discovery Gene Expression – Merck Protein DNA RNA
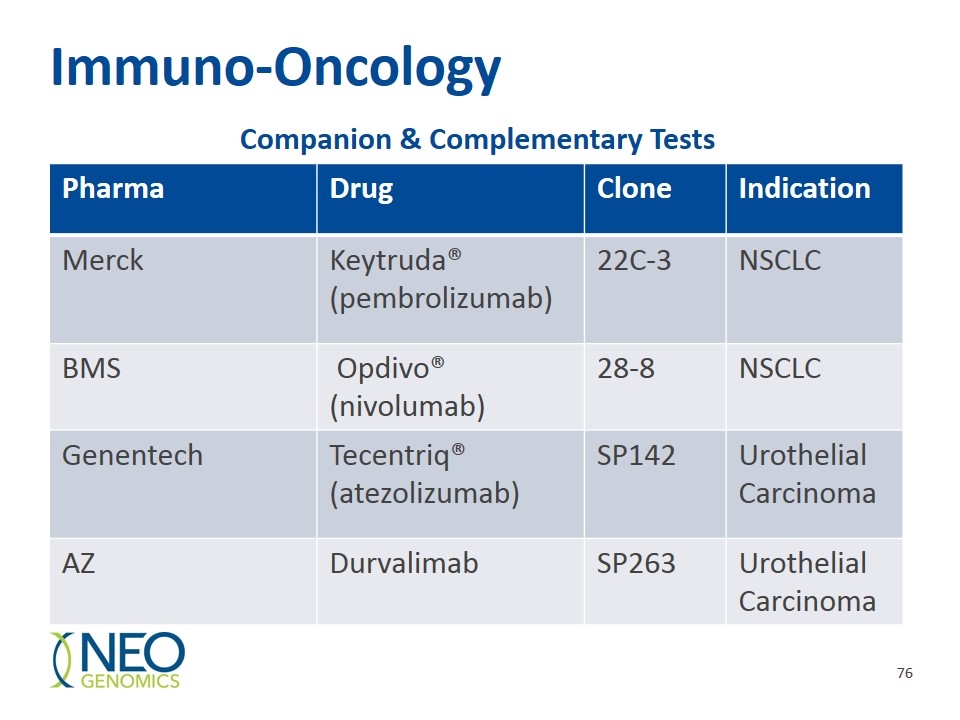
Immuno-Oncology Pharma Drug Clone Indication Merck Keytruda® (pembrolizumab) 22C-3 NSCLC BMS Opdivo® (nivolumab) 28-8 NSCLC Genentech Tecentriq® (atezolizumab) SP142 Urothelial Carcinoma AZ Durvalimab SP263 Urothelial Carcinoma Companion & Complementary Tests
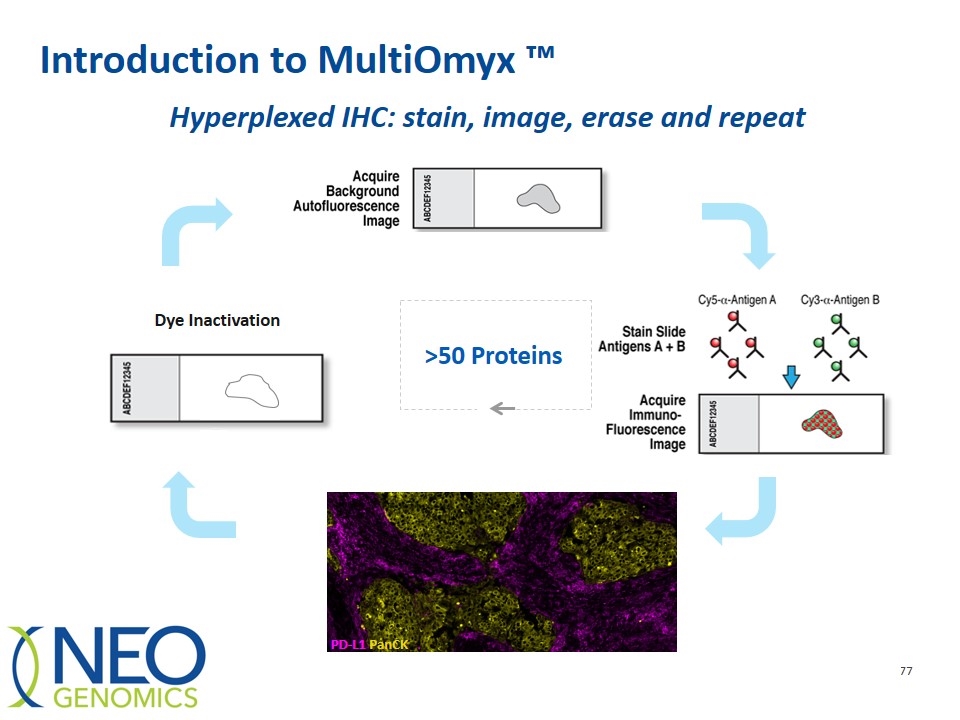
Introduction to MultiOmyx ™ Dye Inactivation >50 Proteins PD-L1 PanCK Hyperplexed IHC: stain, image, erase and repeat
Benefits of MultiOmyx™ for Immunotherapy tests Co-localization of markers can determine the exact phenotype of the cell The only platform that can systematically assess the area around the tumor Quantify beyond the “by eye” ability which can only “bucket” Can systematically look at distance as well as co-expression
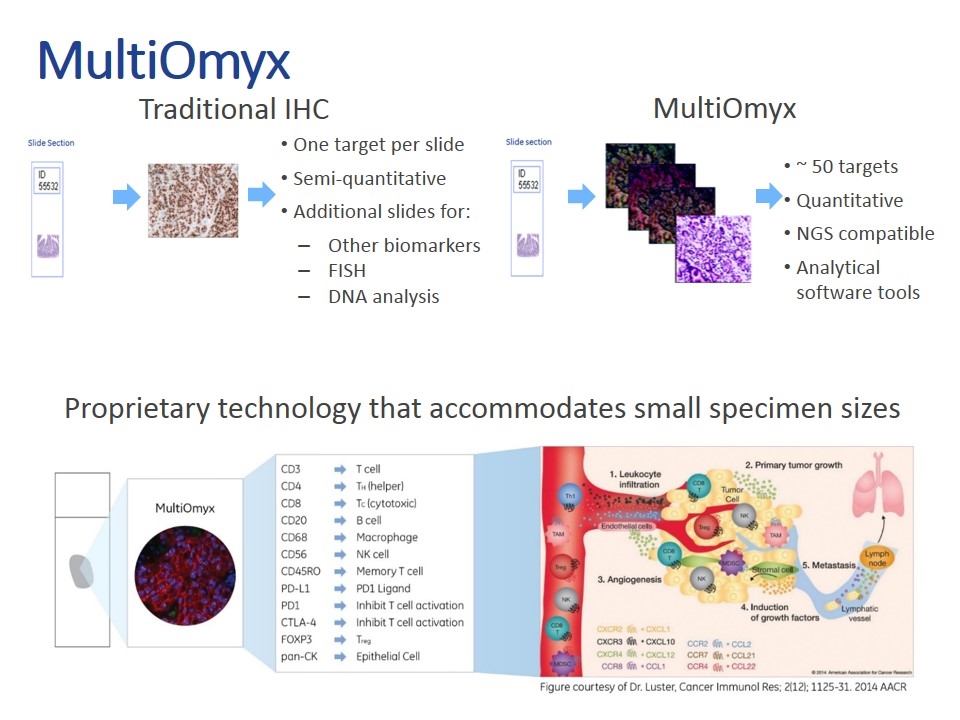
MultiOmyx One target per slide Semi-quantitative Additional slides for: Other biomarkers FISH DNA analysis ~ 50 targets Quantitative NGS compatible Analytical software tools Traditional IHC MultiOmyx Proprietary technology that accommodates small specimen sizes
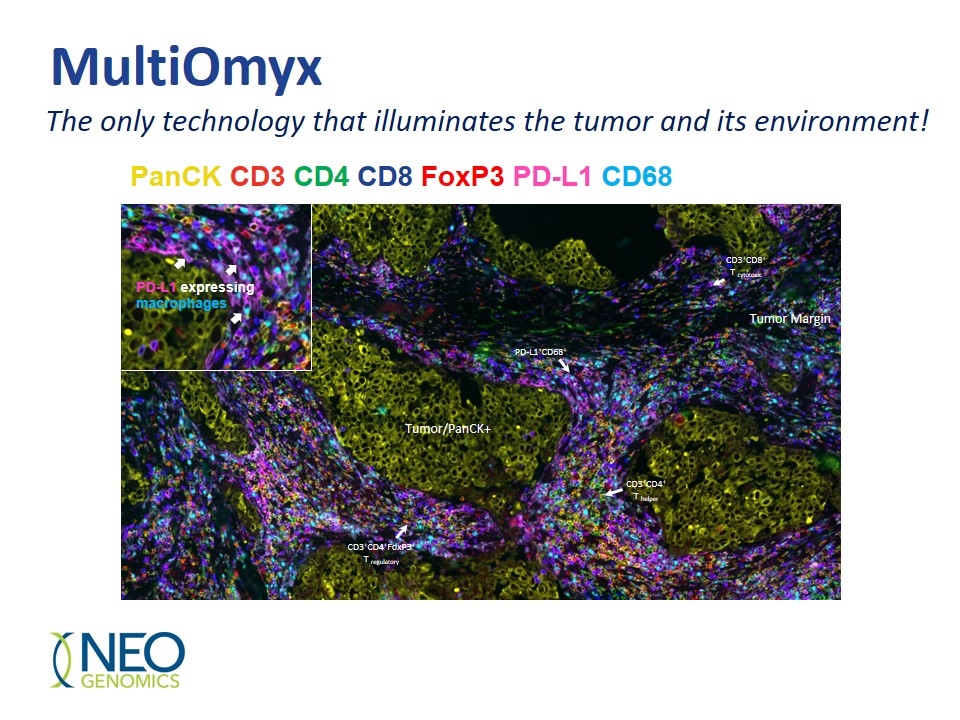
PanCK CD3 CD4 CD8 FoxP3 PD-L1 CD68 The only technology that illuminates the tumor and its environment! Tumor/PanCK+ CD3+CD4+ T helper CD3+CD4+FoxP3+ T regulatory PD-L1+CD68+ CD3+CD8+ T cytotoxic Tumor Margin PD-L1 expressing macrophages MultiOmyx
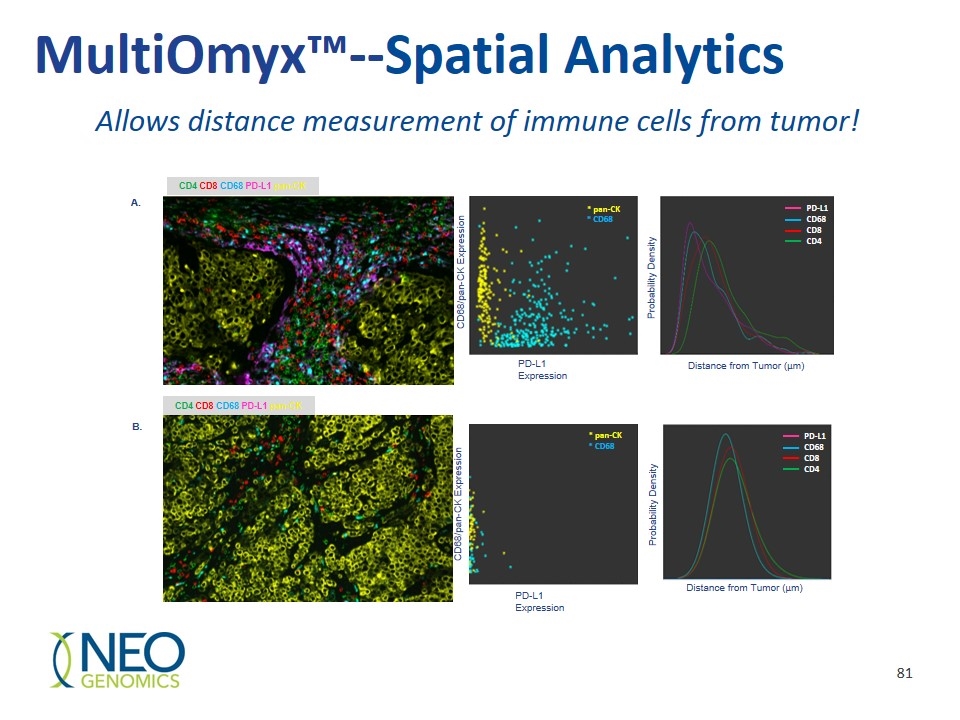
CD4 CD8 CD68 PD-L1 pan-CK * pan-CK * CD68 PD-L1 Expression CD68/pan-CK Expression Distance from Tumor (µm) Distance from Tumor (µm) Probability Density Probability Density PD-L1 CD68 CD8 CD4 CD4 CD8 CD68 PD-L1 pan-CK * pan-CK * CD68 PD-L1 Expression CD68/pan-CK Expression PD-L1 CD68 CD8 CD4 A. B. MultiOmyx™--Spatial Analytics Allows distance measurement of immune cells from tumor!

NeoGenomics Europe, S.A. Route de l’Etraz A1, Rolle Scheduled opening: September 2017 New 740 square meter laboratory Situated midway between Geneva & Lausanne, in the Canton of Vaud Initial on-site capabilities: Flow Cytometry, FISH and IHC based on current Sponsor needs Additional capabilities will be determined by Sponsor needs and timelines 10 year tax holiday Asia laboratory to follow

Key Takeaways þRapidly growing market þWell-positioned in terms of expertise, technology, and growth þInvesting to support growth þImmuno-Oncology has changed the oncology market þMultiOmyx is a key differentiator in the immuno-oncology space

Questions and Answers

R&D and New Test Development Maher Albitar, MD Senior Vice President, Chief Medical Officer and Director of R&D

Maher Albitar, M.D. Senior Vice President, Chief Medical Officer and Director of R&D Dr. Albitar has served as Chief Medical Officer and Director of Research and Development since January 2012. Prior to that Dr. Albitar served as the Medical Director of Hematopathology and Oncology and Chief of R&D at Nichols Institute, Quest Diagnostics. Prior to joining Quest diagnostics, Dr. Albitar was a tenured full professor at MD Anderson Cancer Center and the University of Texas. At MD Anderson, he held the positions of Director of the Molecular Laboratory and Chief of Leukemia in the Division of Pathology and Laboratory Medicine. Dr. Albitar received training in Anatomic and Clinical Pathology at Brown University in Providence, RI and in Hematopathology at the University of Pennsylvania. Dr. Albitar also did post-doctoral training in Genetics at the Genetic Department and Howard Hughes Medical Institute at the University of Pennsylvania. Dr. Albitar received his medical degree in 1979 from Damascus Medical School in Damascus, Syria. Throughout his career, Dr. Albitar has focused on hematologic disease, cancer and molecular biology. He published almost 300 different articles, book chapters and reviews and has multiple patents and patent applications.

Key Drivers of our Leadership in Oncology Testing Expanding Molecular Testing: integrating molecular in Anatomic pathology, Flow cytometry, FISH and cytogenetics Unique molecular capabilities: Proprietary high sensitivity NGS, RNA Sequencing and Quantitative Profiling Leadership in Liquid biopsy (Hematologic neoplasms and prostate cancer) Comprehensive multimodality cancer Genomic profiling and Immunologic profiling Integrating deep learning

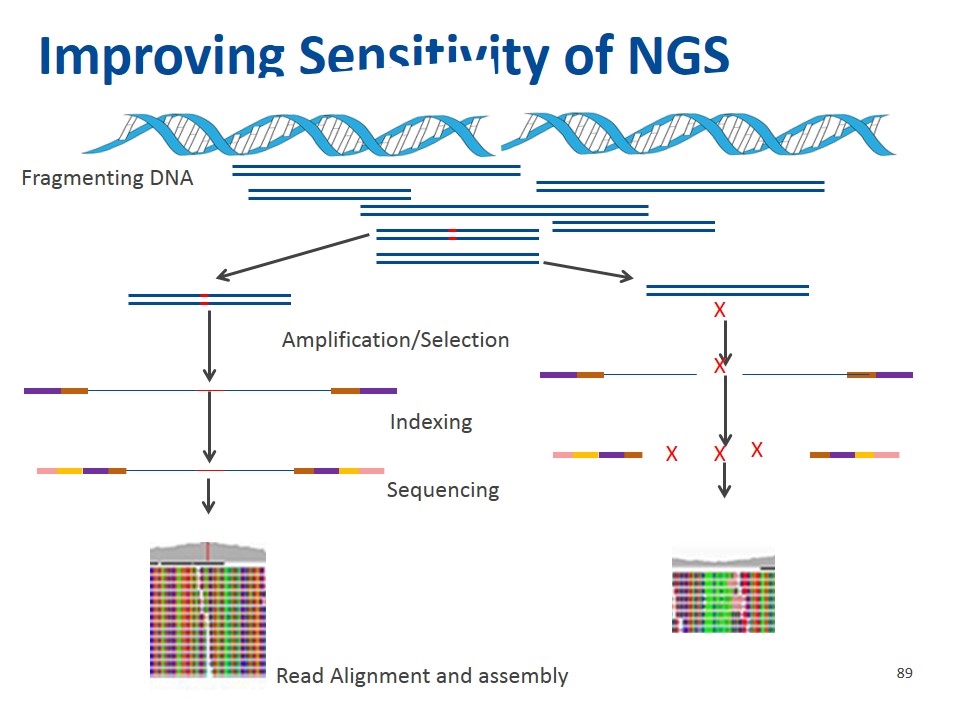
Improving Sensitivity of NGS Read Alignment and assembly Fragmenting DNA Indexing Sequencing Amplification/Selection X X X X X
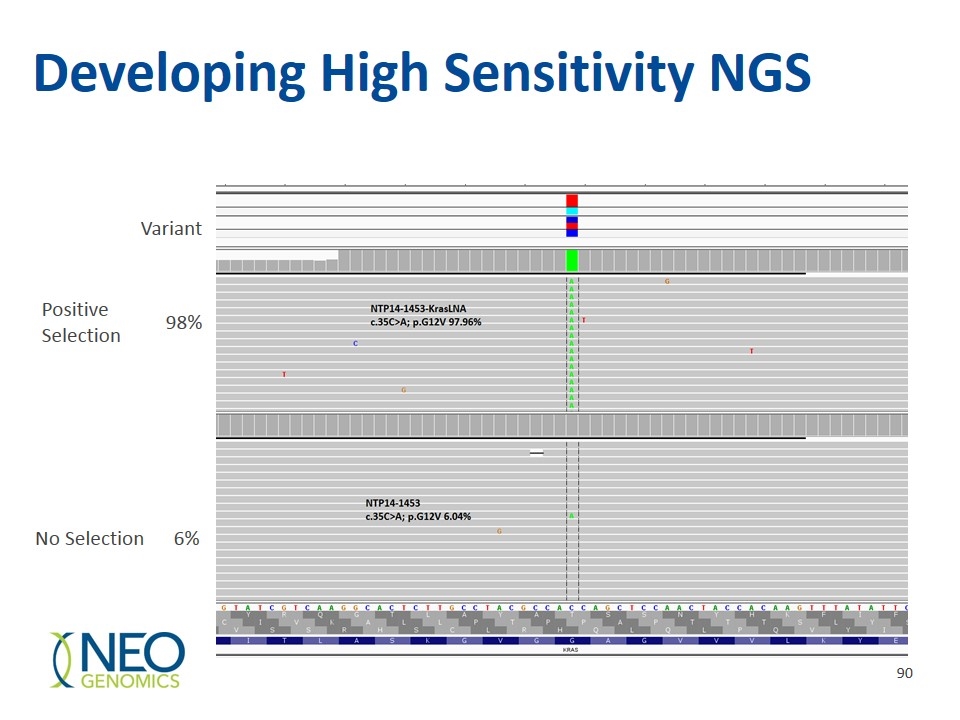
Developing High Sensitivity NGS Positive Selection No Selection 6% 98% Variant
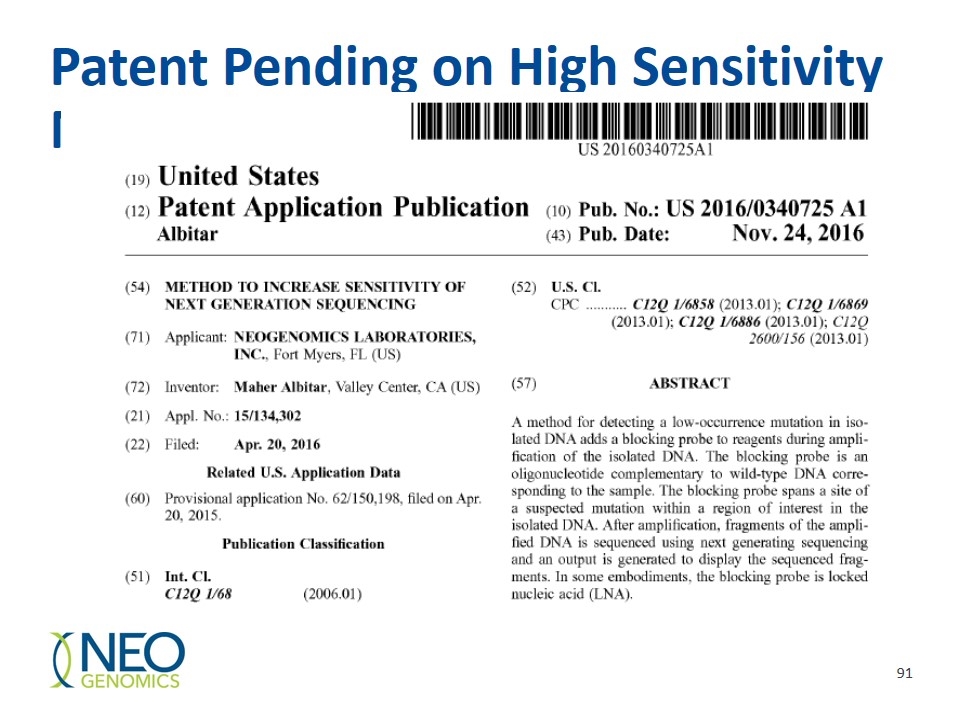
Patent Pending on High Sensitivity NGS
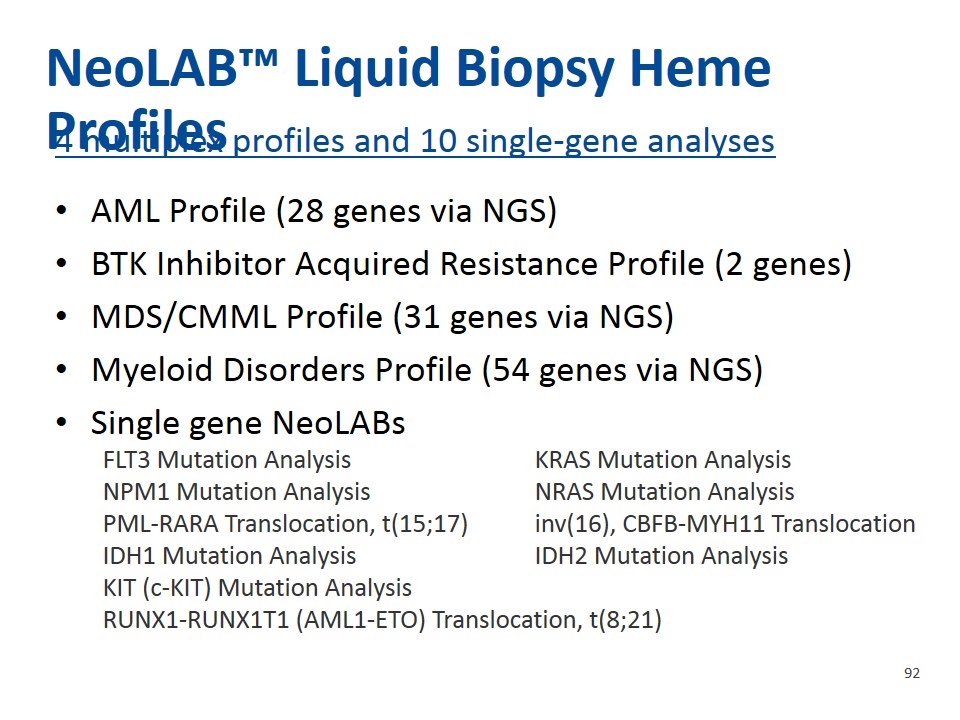
NeoLAB™ Liquid Biopsy Heme Profiles 4 multiplex profiles and 10 single-gene analyses AML Profile (28 genes via NGS) BTK Inhibitor Acquired Resistance Profile (2 genes) MDS/CMML Profile (31 genes via NGS) Myeloid Disorders Profile (54 genes via NGS) Single gene NeoLABs FLT3 Mutation AnalysisKRAS Mutation Analysis NPM1 Mutation AnalysisNRAS Mutation Analysis PML-RARA Translocation, t(15;17)inv(16), CBFB-MYH11 Translocation IDH1 Mutation AnalysisIDH2 Mutation Analysis KIT (c-KIT) Mutation Analysis RUNX1-RUNX1T1 (AML1-ETO) Translocation, t(8;21)
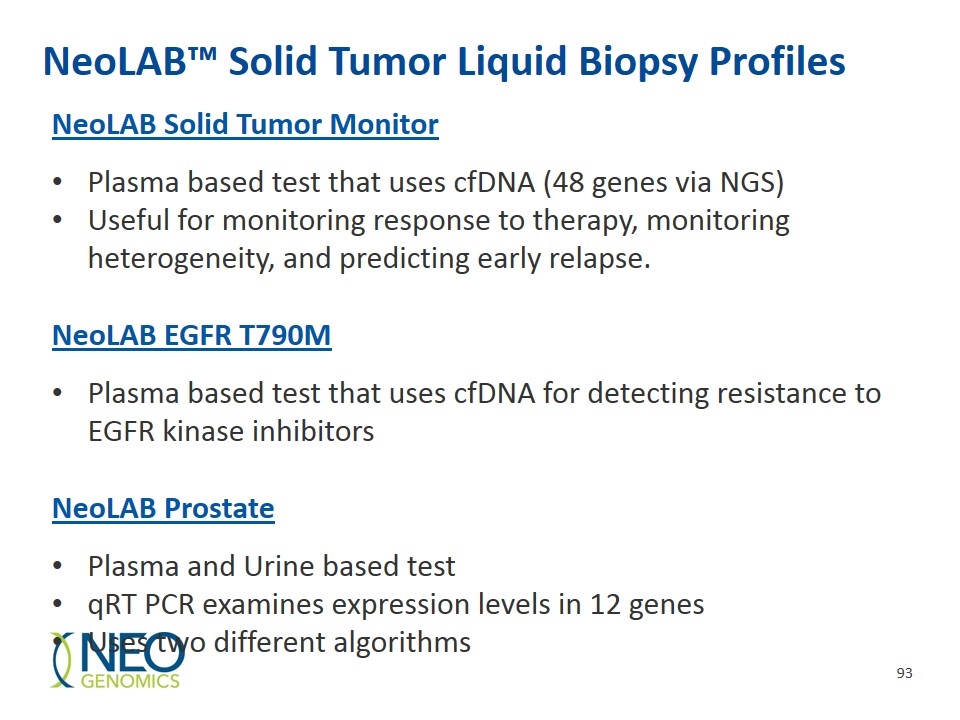
NeoLAB™ Solid Tumor Liquid Biopsy Profiles NeoLAB Solid Tumor Monitor Plasma based test that uses cfDNA (48 genes via NGS) Useful for monitoring response to therapy, monitoring heterogeneity, and predicting early relapse. NeoLAB EGFR T790M Plasma based test that uses cfDNA for detecting resistance to EGFR kinase inhibitors NeoLAB Prostate Plasma and Urine based test qRT PCR examines expression levels in 12 genes Uses two different algorithms
Hematologic Neoplasms Solid Tumors Emerging Leader in Liquid Biopsies
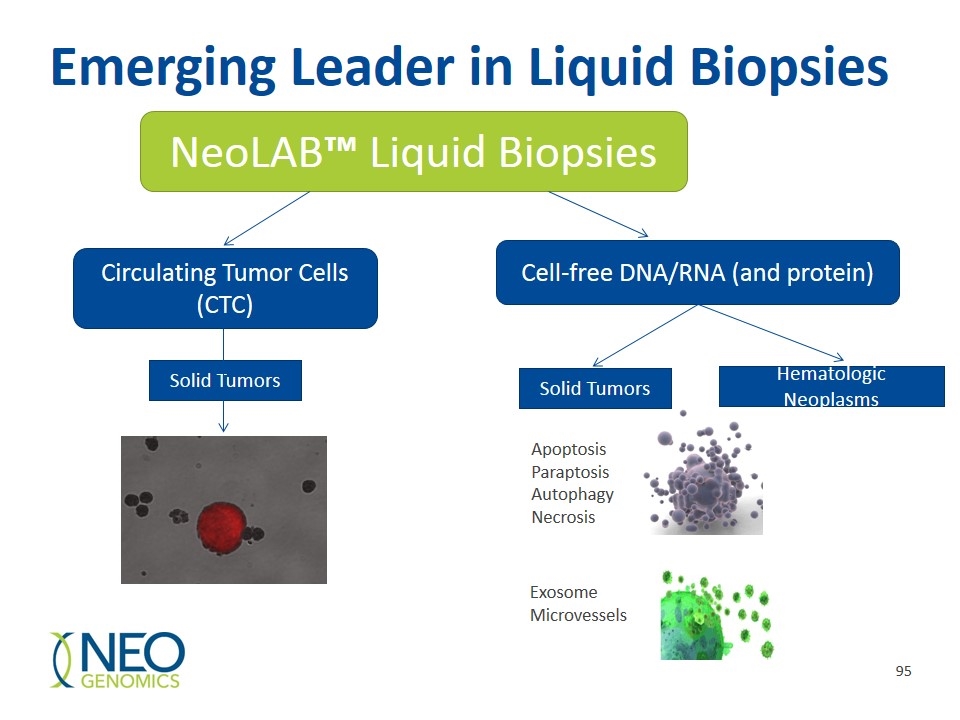
Emerging Leader in Liquid Biopsies NeoLAB™ Liquid Biopsies Circulating Tumor Cells (CTC) Cell-free DNA/RNA (and protein) Solid Tumors Hematologic Neoplasms Solid Tumors Apoptosis Paraptosis Autophagy Necrosis Exosome Microvessels
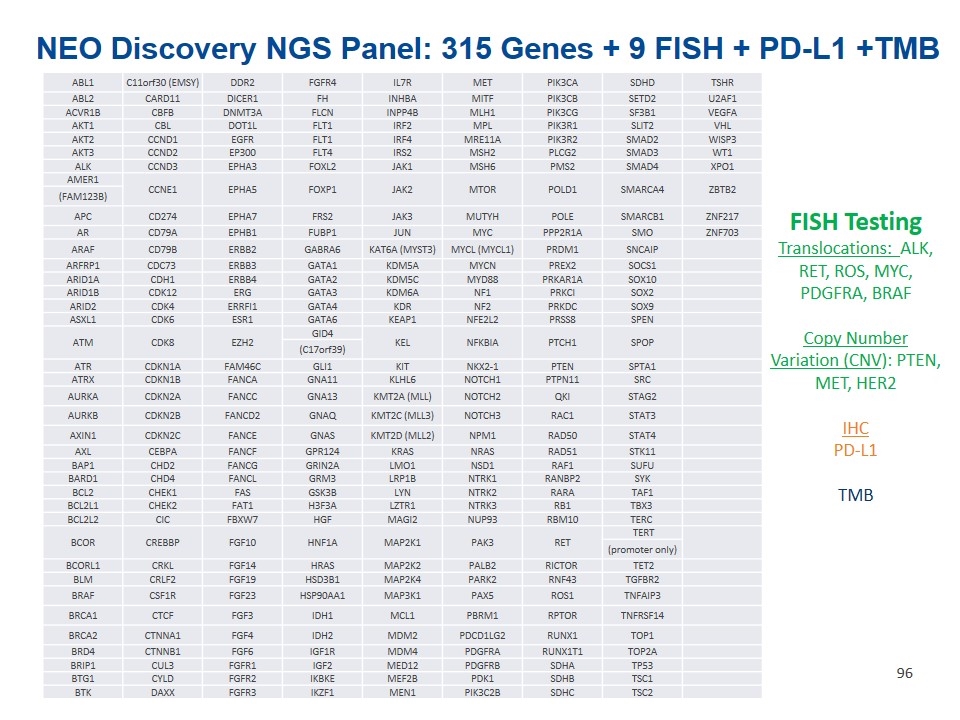
FISH Testing Translocations: ALK, RET, ROS, MYC, PDGFRA, BRAF Copy Number Variation (CNV): PTEN, MET, HER2 IHC PD-L1 TMB ABL1 C11orf30 (EMSY) DDR2 FGFR4 IL7R MET PIK3CA SDHD TSHR ABL2 CARD11 DICER1 FH INHBA MITF PIK3CB SETD2 U2AF1 ACVR1B CBFB DNMT3A FLCN INPP4B MLH1 PIK3CG SF3B1 VEGFA AKT1 CBL DOT1L FLT1 IRF2 MPL PIK3R1 SLIT2 VHL AKT2 CCND1 EGFR FLT1 IRF4 MRE11A PIK3R2 SMAD2 WISP3 AKT3 CCND2 EP300 FLT4 IRS2 MSH2 PLCG2 SMAD3 WT1 ALK CCND3 EPHA3 FOXL2 JAK1 MSH6 PMS2 SMAD4 XPO1 AMER1 CCNE1 EPHA5 FOXP1 JAK2 MTOR POLD1 SMARCA4 ZBTB2 (FAM123B) APC CD274 EPHA7 FRS2 JAK3 MUTYH POLE SMARCB1 ZNF217 AR CD79A EPHB1 FUBP1 JUN MYC PPP2R1A SMO ZNF703 ARAF CD79B ERBB2 GABRA6 KAT6A (MYST3) MYCL (MYCL1) PRDM1 SNCAIP ARFRP1 CDC73 ERBB3 GATA1 KDM5A MYCN PREX2 SOCS1 ARID1A CDH1 ERBB4 GATA2 KDM5C MYD88 PRKAR1A SOX10 ARID1B CDK12 ERG GATA3 KDM6A NF1 PRKCI SOX2 ARID2 CDK4 ERRFI1 GATA4 KDR NF2 PRKDC SOX9 ASXL1 CDK6 ESR1 GATA6 KEAP1 NFE2L2 PRSS8 SPEN ATM CDK8 EZH2 GID4 KEL NFKBIA PTCH1 SPOP (C17orf39) ATR CDKN1A FAM46C GLI1 KIT NKX2-1 PTEN SPTA1 ATRX CDKN1B FANCA GNA11 KLHL6 NOTCH1 PTPN11 SRC AURKA CDKN2A FANCC GNA13 KMT2A (MLL) NOTCH2 QKI STAG2 AURKB CDKN2B FANCD2 GNAQ KMT2C (MLL3) NOTCH3 RAC1 STAT3 AXIN1 CDKN2C FANCE GNAS KMT2D (MLL2) NPM1 RAD50 STAT4 AXL CEBPA FANCF GPR124 KRAS NRAS RAD51 STK11 BAP1 CHD2 FANCG GRIN2A LMO1 NSD1 RAF1 SUFU BARD1 CHD4 FANCL GRM3 LRP1B NTRK1 RANBP2 SYK BCL2 CHEK1 FAS GSK3B LYN NTRK2 RARA TAF1 BCL2L1 CHEK2 FAT1 H3F3A LZTR1 NTRK3 RB1 TBX3 BCL2L2 CIC FBXW7 HGF MAGI2 NUP93 RBM10 TERC BCOR CREBBP FGF10 HNF1A MAP2K1 PAK3 RET TERT (promoter only) BCORL1 CRKL FGF14 HRAS MAP2K2 PALB2 RICTOR TET2 BLM CRLF2 FGF19 HSD3B1 MAP2K4 PARK2 RNF43 TGFBR2 BRAF CSF1R FGF23 HSP90AA1 MAP3K1 PAX5 ROS1 TNFAIP3 BRCA1 CTCF FGF3 IDH1 MCL1 PBRM1 RPTOR TNFRSF14 BRCA2 CTNNA1 FGF4 IDH2 MDM2 PDCD1LG2 RUNX1 TOP1 BRD4 CTNNB1 FGF6 IGF1R MDM4 PDGFRA RUNX1T1 TOP2A BRIP1 CUL3 FGFR1 IGF2 MED12 PDGFRB SDHA TP53 BTG1 CYLD FGFR2 IKBKE MEF2B PDK1 SDHB TSC1 BTK DAXX FGFR3 IKZF1 MEN1 PIK3C2B SDHC TSC2 NEO Discovery NGS Panel: 315 Genes + 9 FISH + PD-L1 +TMB
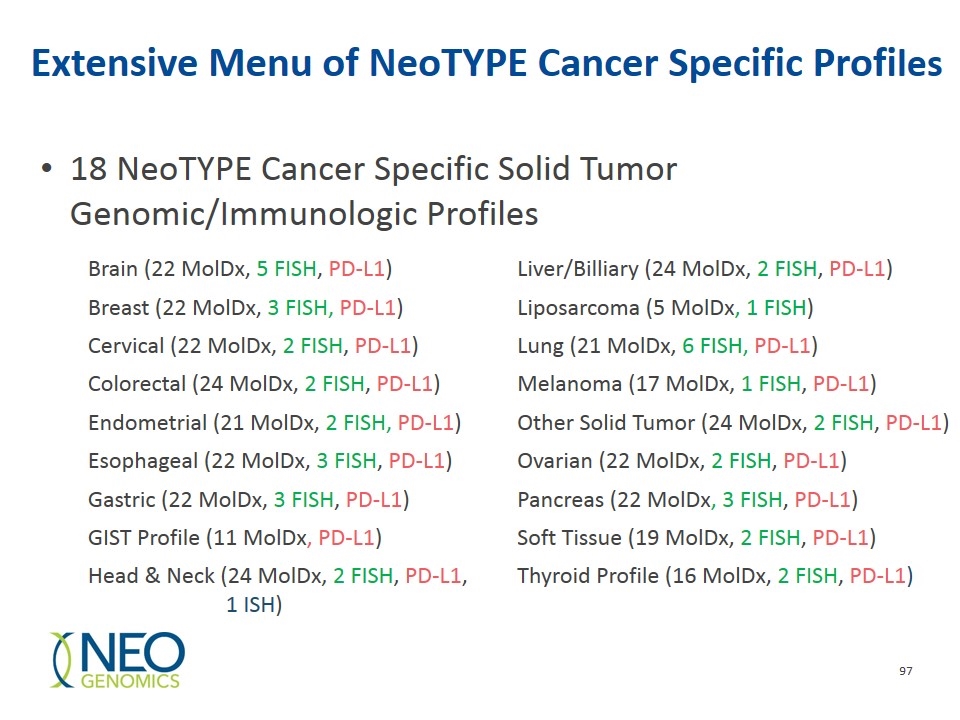
Extensive Menu of NeoTYPE Cancer Specific Profiles Brain (22 MolDx, 5 FISH, PD-L1) Liver/Billiary (24 MolDx, 2 FISH, PD-L1) Breast (22 MolDx, 3 FISH, PD-L1) Liposarcoma (5 MolDx, 1 FISH) Cervical (22 MolDx, 2 FISH, PD-L1) Lung (21 MolDx, 6 FISH, PD-L1) Colorectal (24 MolDx, 2 FISH, PD-L1) Melanoma (17 MolDx, 1 FISH, PD-L1) Endometrial (21 MolDx, 2 FISH, PD-L1) Other Solid Tumor (24 MolDx, 2 FISH, PD-L1) Esophageal (22 MolDx, 3 FISH, PD-L1) Ovarian (22 MolDx, 2 FISH, PD-L1) Gastric (22 MolDx, 3 FISH, PD-L1) Pancreas (22 MolDx, 3 FISH, PD-L1) GIST Profile (11 MolDx, PD-L1) Soft Tissue (19 MolDx, 2 FISH, PD-L1) Head & Neck (24 MolDx, 2 FISH, PD-L1, 1 ISH) Thyroid Profile (16 MolDx, 2 FISH, PD-L1) 18 NeoTYPE Cancer Specific Solid Tumor Genomic/Immunologic Profiles
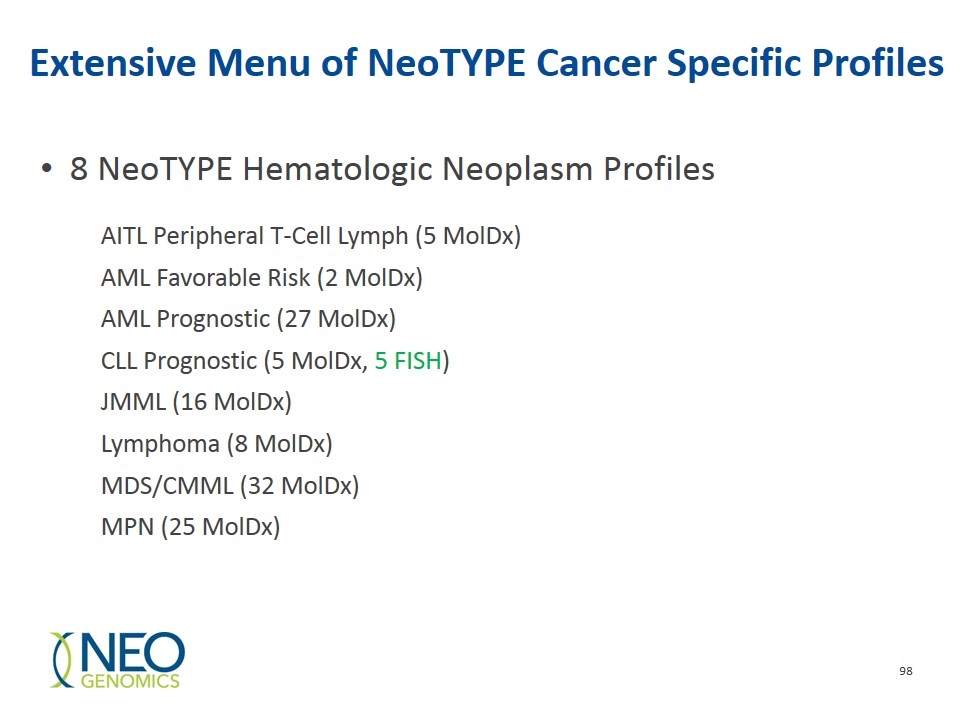
Extensive Menu of NeoTYPE Cancer Specific Profiles AITL Peripheral T-Cell Lymph (5 MolDx) AML Favorable Risk (2 MolDx) AML Prognostic (27 MolDx) CLL Prognostic (5 MolDx, 5 FISH) JMML (16 MolDx) Lymphoma (8 MolDx) MDS/CMML (32 MolDx) MPN (25 MolDx) 8 NeoTYPE Hematologic Neoplasm Profiles
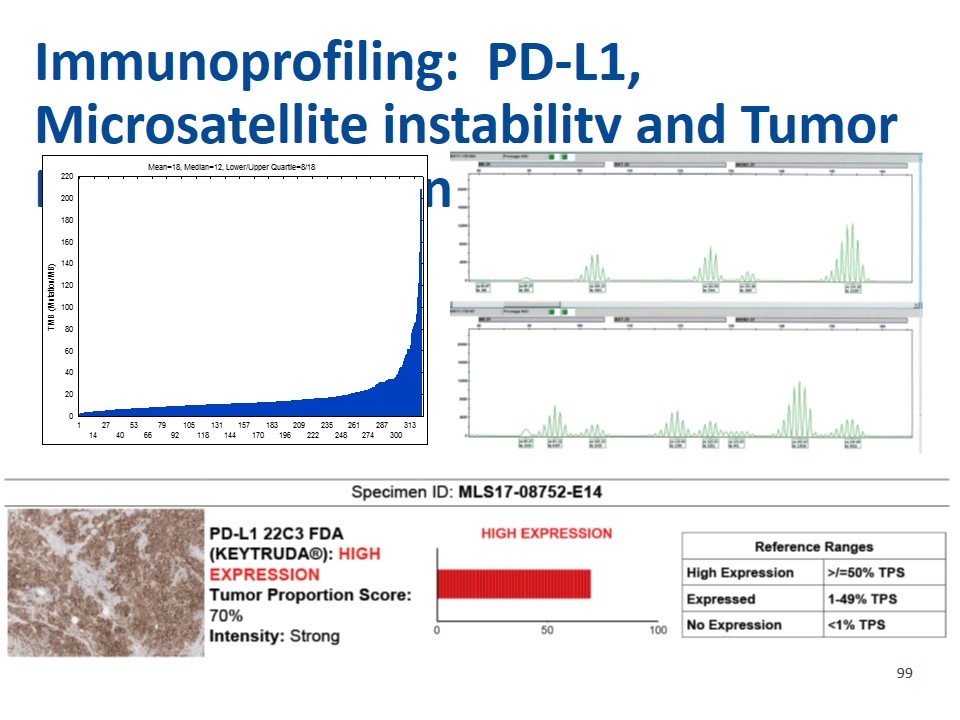
Immunoprofiling: PD-L1, Microsatellite instability and Tumor Mutation Burden (TMB)
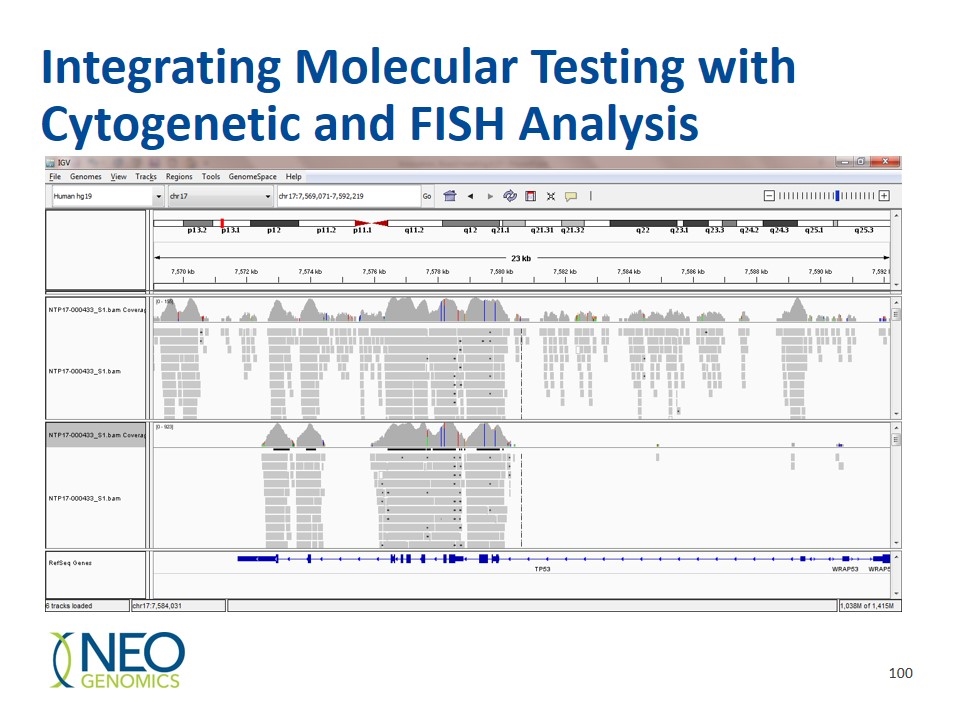
Integrating Molecular Testing with Cytogenetic and FISH Analysis
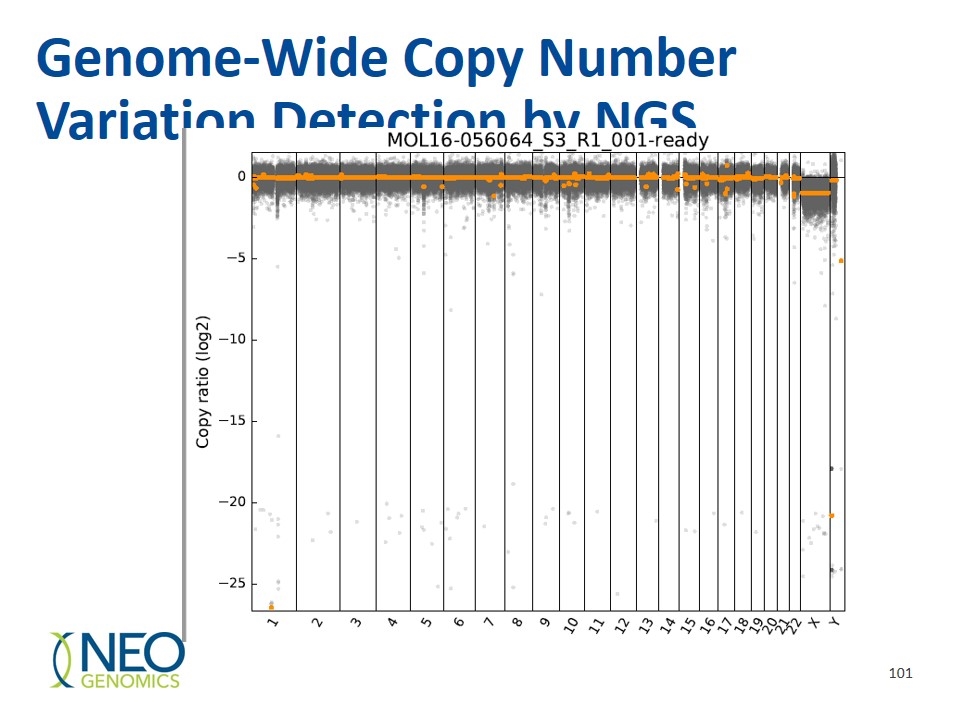
Genome-Wide Copy Number Variation Detection by NGS
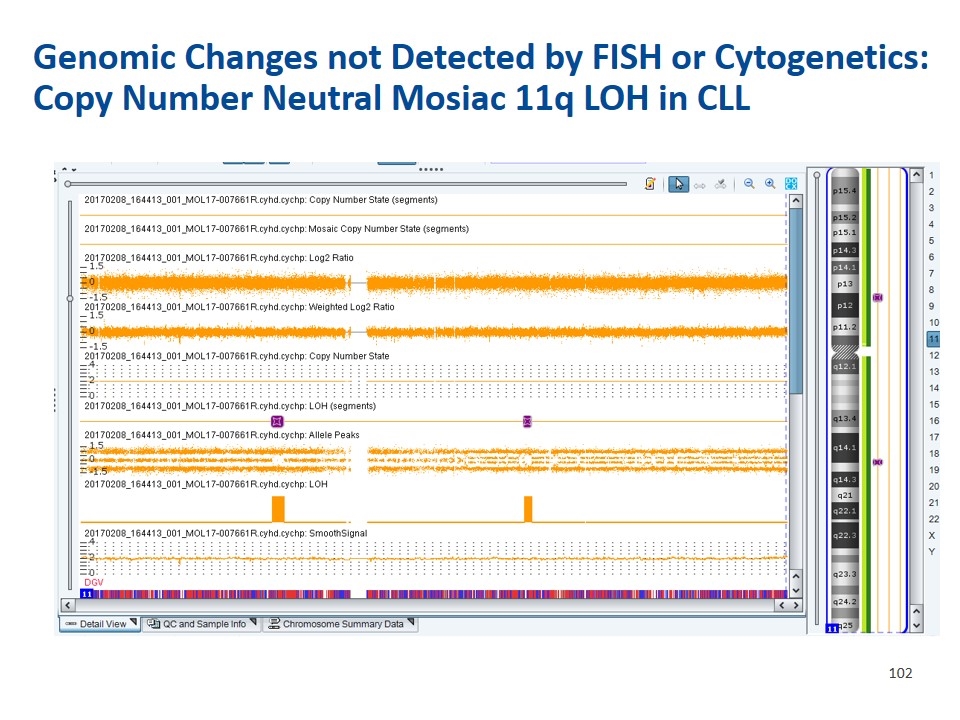
Genomic Changes not Detected by FISH or Cytogenetics: Copy Number Neutral Mosiac 11q LOH in CLL
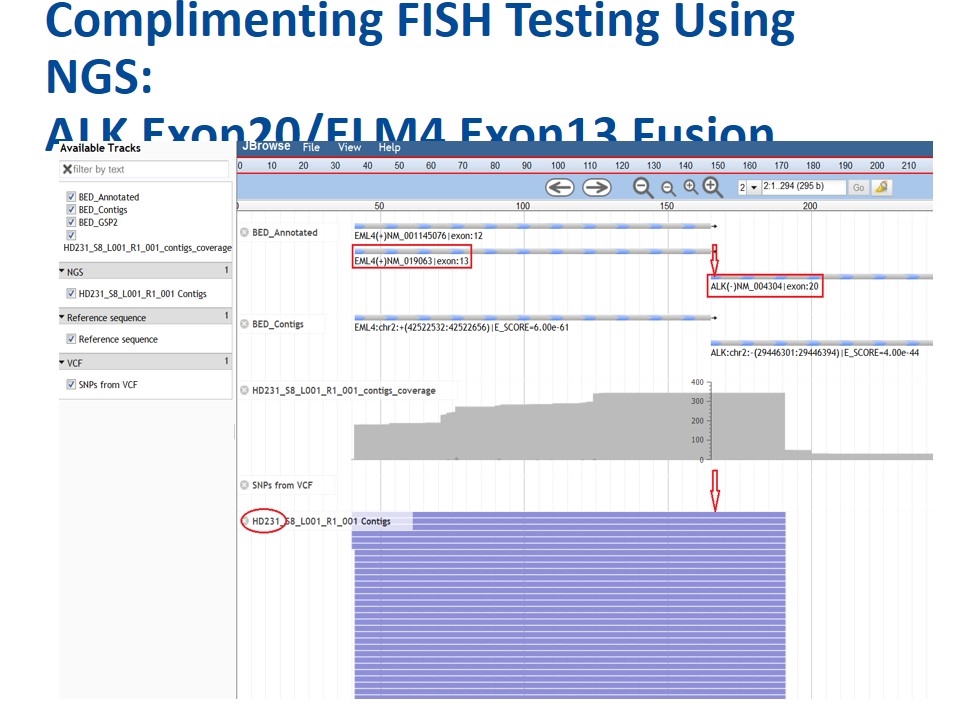
Complimenting FISH Testing Using NGS: ALK Exon20/ELM4 Exon13 Fusion
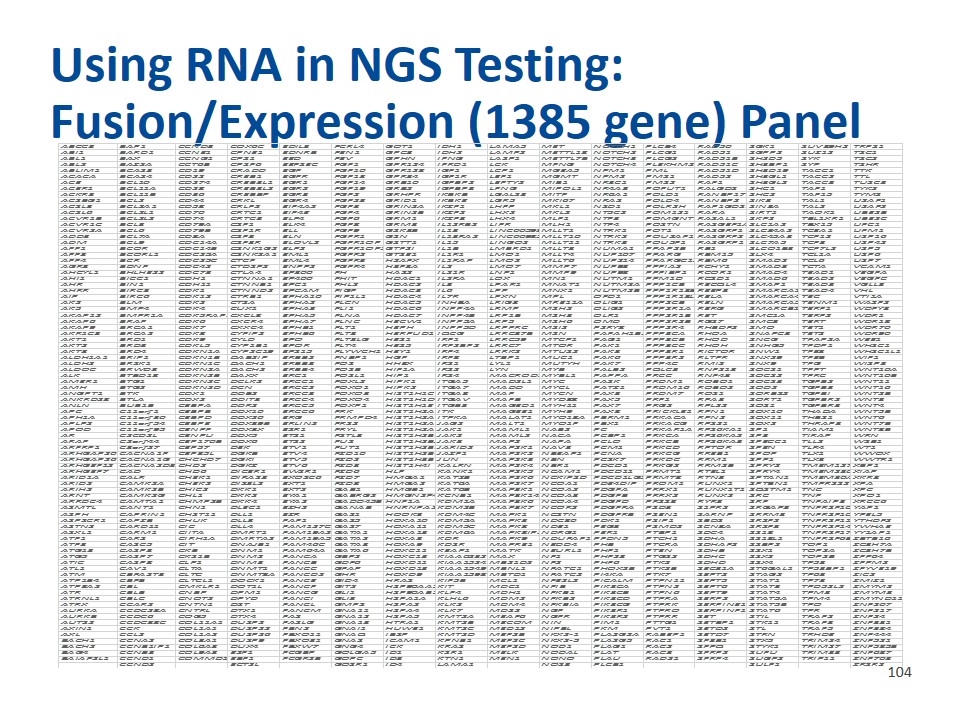
Using RNA in NGS Testing: Fusion/Expression (1385 gene) Panel ABCC3 BAP1 CCND3 COX6C EDIL3 FCRL4 GOT1 IDH1 LAMA5 MET NOTCH1 PLCB4 RAD50 SGK1 SUV39H2 TRPS1 ABI1 BARD1 CCNE1 CPNE1 EDNRB FEN1 GPC3 IDH2 LAMP2 METTL18 NOTCH2 PLCG1 RAD51 SGPP2 SUZ12 TSC1 ABL1 BAX CCNG1 CPS1 EED FEV GPHN IFNG LASP1 METTL7B NOTCH3 PLCG2 RAD51B SH2D5 SYK TSC2 ABL2 BAZ2A CCT6B CPSF6 EEFSEC FGF1 GPR124 IFRD1 LCK MFNG NOTCH4 PLEKHM2 RAD51C SH3BP1 SYP TSHR ABLIM1 BCAS3 CD19 CRADD EGF FGF10 GPR128 IGF1 LCP1 MGEA5 NPM1 PML RAD51D SH3D19 TACC1 TTK ACACA BCAS4 CD22 CREB1 EGFR FGF13 GPR34 IGF1R LEF1 MGMT NPM2 PMS1 RAD52 SH3GL1 TACC2 TTL ACE BCL10 CD274 CREB3L1 EGR1 FGF14 GRB10 IGFBP2 LEFTY2 MIB1 NR3C1 PMS2 RAF1 SH3GL2 TACC3 TUSC3 ACER1 BCL11A CD28 CREB3L2 EGR2 FGF19 GRB2 IGFBP3 LFNG MIPOL1 NR4A3 POFUT1 RALGDS SHC1 TAF1 TYK2 ACKR3 BCL11B CD36 CREBBP EGR3 FGF2 GRHPR IKBKB LGALS3 MITF NR6A1 POLD1 RANBP17 SHC2 TAF15 TYMS ACSBG1 BCL2 CD44 CRKL EGR4 FGF23 GRID1 IKBKE LGR5 MKI67 NRAS POLD4 RANBP2 SIK3 TAL1 U2AF1 ACSL3 BCL2A1 CD58 CRLF2 EIF4A2 FGF3 GRIN2A IKZF1 LHFP MKL1 NSD1 POLR2H RAP1GDS1 SIN3A TAL2 U2AF2 ACSL6 BCL2L1 CD70 CRTC1 EIF4E FGF4 GRIN2B IKZF2 LHX2 MKL2 NT5C2 POM121 RARA SIRT1 TAOK1 UBE2B ACVR1B BCL2L2 CD74 CRTC3 ELF4 FGF6 GRM1 IKZF3 LHX4 MLF1 NTF3 POMGNT1 RASAL1 SKP2 TBL1XR1 UBE2C ACVR1C BCL3 CD79A CSF1 ELK4 FGF8 GRM3 IL12RB2 LIFR MLH1 NTF4 POSTN RASGEF1A SLC1A2 TBX15 UFC1 ACVR2A BCL6 CD79B CSF1R ELL FGF9 GSK3B IL13 LINC00598 MLLT1 NTRK1 POT1 RASGRF1 SLC34A2 TCEA1 UFM1 ADD3 BCL7A CD8A CSF3 ELN FGFR1 GSN IL13RA2 LINC00982 MLLT10 NTRK2 POU2AF1 RASGRF2 SLC45A3 TCF12 USP16 ADM BCL9 CDC14A CSF3R ELOVL2 FGFR1OP GSTT1 IL15 LINGO2 MLLT11 NTRK3 POU5F1 RASGRP1 SLC7A5 TCF3 USP42 AFF1 BCOR CDC14B CSNK1G2 ELP2 FGFR1OP2 GTF2I IL1B LMBRD1 MLLT3 NUMA1 PPAP2B RB1 SLCO1B3 TCF7L2 USP5 AFF3 BCORL1 CDC25A CSNK2A1 EML1 FGFR2 GTSE1 IL1R1 LMO1 MLLT4 NUP107 PPARG RBM15 SLX4 TCL1A USP6 AFF4 BCR CDC25C CTCF EML4 FGFR3 H2AFX IL1RAP LMO2 MLLT6 NUP214 PPARGC1A RBM6 SMAD2 TCL6 USP7 AGR3 BDNF CDC42 CTDSP2 ENPP2 FGFR4 H3F3A IL2 LMO7 MMP7 NUP93 PPFIA2 RCHY1 SMAD3 TCTA VCAM1 AHCYL1 BHLHE22 CDC73 CTLA4 EP300 FH HAS2 IL21R LNP1 MMP9 NUP98 PPFIBP1 RCOR1 SMAD4 TEAD1 VEGFA AHI1 BICC1 CDH1 CTNNA1 EP400 FHIT HDAC1 IL2RA LOX MN1 NUTM1 PPM1D RCSD1 SMAD6 TEAD2 VEGFC AHR BIN1 CDH11 CTNNB1 EPC1 FHL2 HDAC2 IL3 LPAR1 MNAT1 NUTM2A PPP1CB RECQL4 SMAP1 TEAD3 VGLL3 AHRR BIRC3 CDK1 CTNND2 EPCAM FIGF HDAC3 IL6 LPP MNX1 NUTM2B PPP1R13B REEP3 SMARCA1 TEAD4 VHL AIP BIRC6 CDK12 CTRB1 EPHA10 FIP1L1 HDAC4 IL7R LPXN MPL OFD1 PPP1R13L RELA SMARCA4 TEC VTI1A AK2 BLM CDK2 CTSA EPHA2 FLCN HDAC5 INHBA LRIG3 MRE11A OLIG1 PPP2CB RELN SMARCA5 TENM1 WASF2 AK5 BMP4 CDK4 CUX1 EPHA3 FLI1 HDAC6 INPP4A LRMP MSH2 OLIG2 PPP2R1A RERG SMARCB1 TERF1 WDFY3 AKAP12 BMPR1A CDK5RAP2 CXCL8 EPHA5 FLNA HDAC7 INPP4B LRP1B MSH3 OLR1 PPP2R1B RET SMC1A TERF2 WDR1 AKAP6 BRAF CDK6 CXCR4 EPHA7 FLNC HECW1 INPP5A LRP5 MSH6 OMD PPP2R2B RGS7 SMC3 TERT WDR18 AKAP9 BRCA1 CDK7 CXXC4 EPHB1 FLT1 HEPH INPP5D LRPPRC MSI2 P2RY8 PPP2R4 RHBDF2 SMO TET1 WDR70 AKR1C3 BRCA2 CDK8 CYFIP2 EPHB6 FLT3 HERPUD1 IQCG LRRC37B MSN PAFAH1B2 PPP3CA RHOA SNAPC3 TET2 WDR90 AKT1 BRD1 CDK9 CYLD EPO FLT3LG HES1 IRF1 LRRC59 MTCP1 PAG1 PPP3CB RHOD SNCG TFAP2A WEE1 AKT2 BRD3 CDKL5 CYP1B1 EPOR FLT4 HES5 IRF2BP2 LRRC7 MTOR PAK1 PPP3CC RHOH SNHG5 TFDP1 WHSC1 AKT3 BRD4 CDKN1A CYP2C19 EPS15 FLYWCH1 HEY1 IRF4 LRRK2 MTUS2 PAK3 PPP3R1 RICTOR SNW1 TFE3 WHSC1L1 ALDH1A1 BRIP1 CDKN1B DAB2IP ERBB2 FNBP1 HGF IRF8 LTBP1 MUC1 PAK6 PPP3R2 RLTPR SNX29 TFEB WIF1 ALDH2 BRSK1 CDKN1C DACH1 ERBB3 FOS HHEX IRS1 LYL1 MUTYH PAK7 PPP4C RMI2 SNX9 TFG WISP3 ALDOC BRWD3 CDKN2A DACH2 ERBB4 FOSB HIF1A IRS2 LYN MYB PALB2 PQLC3 RNF213 SOCS1 TFPT WNT10A ALK BTBD18 CDKN2B DAXX ERC1 FOSL1 HIP1 IRS4 MACROD1 MYBL1 PAPPA PRCC RNF43 SOCS2 TFRC WNT10B AMER1 BTG1 CDKN2C DCLK2 ERCC1 FOXL2 HIPK1 ITGA5 MAD2L1 MYC PASK PRDM1 ROBO1 SOCS3 TGFB2 WNT11 AMH BTG2 CDKN2D DCN ERCC2 FOXO1 HIPK2 ITGA7 MADD MYCL PATZ1 PRDM16 ROBO2 SOD2 TGFB3 WNT16 ANGPT1 BTK CDX1 DDB2 ERCC3 FOXO3 HIST1H1C ITGA8 MAF MYCN PAX3 PRDM7 ROS1 SORBS2 TGFBI WNT2B ANKRD28 BTLA CDX2 DDIT3 ERCC4 FOXO4 HIST1H1D ITGAV MAFB MYD88 PAX5 PRF1 RPA3 SORT1 TGFBR2 WNT3 ANLN BUB1B CEBPA DDR2 ERCC5 FOXP1 HIST1H1E ITGB3 MAGED1 MYH11 PAX7 PRG2 RPL22 SOS1 TGFBR3 WNT4 APC C11orf1 CEBPB DDX10 ERCC6 FRK HIST1H2AC ITK MAGEE1 MYH9 PAX8 PRICKLE1 RPN1 SOX10 THADA WNT5B APH1A C11orf30 CEBPD DDX20 ERG FRMPD4 HIST1H2AG ITPKA MALAT1 MYO18A PBRM1 PRKACA RPN2 SOX11 THBS1 WNT6 APLP2 C11orf54 CEBPE DDX39B ERLIN2 FRS2 HIST1H2AL JAG2 MALT1 MYO1F PBX1 PRKACG RPS21 SOX2 THRAP3 WNT7B APOD C11orf95 CENPF DDX3X ESR1 FRYL HIST1H2AM JAK1 MAML1 NAB2 PC PRKAR1A RPS6KA1 SP1 TIAM1 WNT8B AR C2CD2L CENPU DDX5 ETS1 FSTL3 HIST1H2BC JAK2 MAML2 NACA PCBP1 PRKCA RPS6KA2 SP3 TIRAP WRN ARAF C2orf44 CEP170B DDX6 ETS2 FUS HIST1H2BJ JAK3 MAP2 NAPA PCLO PRKCB RPS6KA3 SPECC1 TLL2 WSB1 ARFRP1 C3orf27 CEP57 DEK ETV1 FUT1 HIST1H2BK JARID2 MAP2K1 NAV3 PCM1 PRKCD RPTOR SPEN TLR4 WT1 ARHGAP20 CACNA1F CEP85L DGKB ETV4 FZD10 HIST1H2BO JAZF1 MAP2K2 NBEAP1 PCNA PRKCG RREB1 SPOP TLX1 WWOX ARHGAP26 CACNA1G CHCHD7 DGKI ETV5 FZD2 HIST1H3B JUN MAP2K3 NBN PCSK7 PRKDC RRM1 SPP1 TLX3 WWTR1 ARHGEF12 CACNA2D3 CHD2 DGKZ ETV6 FZD3 HIST1H4I KALRN MAP2K4 NBR1 PDCD1 PRKG2 RRM2B SPRY2 TMEM127 XBP1 ARHGEF7 CAD CHD6 DICER1 EWSR1 FZD6 HLF KANK1 MAP2K5 NCAM1 PDCD11 PRMT1 RTEL1 SPRY4 TMEM230 XIAP ARID1A CALR CHEK1 DIRAS3 EXOSC6 FZD7 HMGA1 KAT2B MAP2K6 NCKIPSD PDCD1LG2 PRMT8 RTN3 SPTAN1 TMEM30A XKR3 ARID2 CAMK2A CHEK2 DIS3L2 EXT1 FZD8 HMGA2 KAT6A MAP2K7 NCOA1 PDE4DIP PROM1 RUNX1 SPTBN1 TMPRSS2 XPA ARIH2 CAMK2B CHIC2 DKK1 EXT2 GAB1 HMGB1 KAT6B MAP3K1 NCOA2 PDGFA PRRX1 RUNX1T1 SQSTM1 TNC XPC ARNT CAMK2G CHL1 DKK2 EYA1 GABRG2 HMGN2P46 KCNB1 MAP3K14 NCOA3 PDGFB PRRX2 RUNX2 SRC TNF XPO1 ARRDC4 CAMTA1 CHMP2B DKK4 EYA2 GADD45B HNF1A KDM1A MAP3K6 NCOA4 PDGFD PRSS8 RYR3 SRF TNFAIP3 XRCC6 ASMTL CANT1 CHN1 DLEC1 EZH2 GANAB HNRNPA2B1 KDM2B MAP3K7 NCOR2 PDGFRA PSD3 S1PR2 SRGAP3 TNFRSF10B YAP1 ASPH CAPRIN1 CHST11 DLL1 EZR GAS1 HOOK3 KDM4C MAPK1 NCSTN PDGFRB PSEN1 SARNP SRRM3 TNFRSF10D YPEL5 ASPSCR1 CAPZB CHUK DLL3 FAF1 GAS5 HOXA10 KDM5A MAPK3 NDC80 PDK1 PSIP1 SBDS SRSF2 TNFRSF11A YTHDF2 ASTN2 CARD11 CIC DLL4 FAM127C GAS7 HOXA11 KDM5C MAPK8 NDE1 PEG3 PSMD2 SCN8A SRSF3 TNFRSF14 YWHAE ASXL1 CARM1 CIITA DMRT1 FAM19A2 GATA1 HOXA13 KDM6A MAPK8IP2 NDRG1 PER1 PTBP1 SDC4 SS18 TNFRSF17 YY1AP1 ATF1 CARS CIRH1A DMRTA2 FAM19A5 GATA2 HOXA3 KDR MAPK9 NDUFAF1 PFDN5 PTCH1 SDHA SS18L1 TNFRSF6B ZBTB16 ATF3 CASC5 CIT DNAJB1 FAM46C GATA3 HOXA9 KDSR MAPRE1 NEDD4 PHB PTCRA SDHAF2 SSBP2 TOP1 ZC3H7A ATG13 CASP3 CKB DNM1 FAM64A GATA6 HOXC11 KEAP1 MATK NEURL1 PHF1 PTEN SDHB SSX1 TOP2A ZC3H7B ATG5 CASP7 CKS1B DNM2 FANCA GBP2 HOXC13 KIAA0232 MAX NF1 PHF23 PTGS2 SDHC SSX2 TOP2B ZFP64 ATIC CASP8 CLP1 DNM3 FANCB GDF6 HOXD11 KIAA1524 MB21D2 NF2 PHF6 PTK2 SDHD SSX4 TP53 ZFPM2 ATL1 CAV1 CLTA DNMT1 FANCC GFAP HOXD13 KIAA1549 MBNL1 NFATC1 PHOX2B PTK2B SEC31A ST6GAL1 TP53BP1 ZFYVE19 ATM CBFA2T3 CLTC DNMT3A FANCD2 GHR HOXD9 KIAA1598 MBTD1 NFATC2 PI4KA PTK7 SEPT2 STAG2 TP63 ZIC2 ATP1B4 CBFB CLTCL1 DOCK1 FANCE GID4 HRAS KIF5B MCL1 NFE2L2 PICALM PTPN11 SEPT5 STAT1 TP73 ZMIZ1 ATP8A2 CBL CMKLR1 DOT1L FANCF GIT2 HSP90AA1 KIT MDC1 NFIB PIK3CA PTPN2 SEPT6 STAT3 TPD52L2 ZMYM2 ATR CBLB CNBP DPM1 FANCG GLI1 HSP90AB1 KLF4 MDH1 NFKB1 PIK3CB PTPN6 SEPT9 STAT4 TPM3 ZMYM3 ATRNL1 CBLC CNOT2 DPYD FANCI GLI3 HSPA1A KLHL6 MDM2 NFKB2 PIK3CD PTPRA SERP2 STAT5A TPM4 ZMYND11 ATRX CCAR2 CNTN1 DST FANCL GMPS HSPA2 KLK2 MDM4 NFKBIA PIK3CG PTPRK SERPINE1 STAT5B TPO ZNF207 AURKA CCDC28A CNTRL DTX1 FANCM GNA11 HSPA4 KLK7 MDS2 NGF PIK3R1 PTPRO SERPINF1 STAT6 TPR ZNF217 AURKB CCDC6 COG5 DTX4 FAS GNA12 HSPA5 KMT2A MEAF6 NGFR PIK3R2 PTPRR SET STIL TRAF2 ZNF24 AUTS2 CCDC88C COL11A1 DUSP2 FASLG GNA13 HTRA1 KMT2B MECOM NIN PIM1 PTTG1 SETBP1 STK11 TRAF3 ZNF331 AXIN1 CCK COL1A1 DUSP22 FBN2 GNAI1 HUWE1 KMT2C MED12 NIPBL PKM PVT1 SETD2 STL TRAF5 ZNF384 AXL CCL2 COL1A2 DUSP26 FBXO11 GNAQ IBSP KMT2D MEF2B NKX2-1 PLA2G2A RABEP1 SETD7 STRN TRHDE ZNF444 BACH1 CCNA2 COL3A1 DUSP9 FBXO31 GNAS ICAM1 KPNB1 MEF2C NKX2-5 PLA2G5 RAC1 SF3B1 STX5 TRIM24 ZNF521 BACH2 CCNB1IP1 COL6A3 DUX4 FBXW7 GNG4 ICK KRAS MEF2D NOD1 PLAG1 RAC2 SFPQ STYK1 TRIM27 ZNF585B BAG4 CCNB3 COL9A3 E2F1 FCGBP GOLGA5 ID1 KSR1 MELK NODAL PLAT RAC3 SFRP2 SUFU TRIM33 ZNF687 BAIAP2L1 CCND1 COMMD1 EBF1 FCGR2B GOPC ID3 KTN1 MEN1 NONO PLAU RAD21 SFRP4 SUGP2 TRIP11 ZNF703 CCND2 ECT2L GOSR1 ID4 LAMA1 NOS3 PLCB1 SULF1 ZRSR2 Some of the genomic variants, genes, nucleic acid sequences, or genomic regions on this list, and their use in specific applications, may be protected by patents. Customers are advised to determine whether they are required to obtain licenses from the party that owns or controls such patents in order to use the product in Customer's specific application. Unless expressly stated otherwise in writing by Illumina, Customer, and not Illumina, is solely responsible for obtaining such licenses.
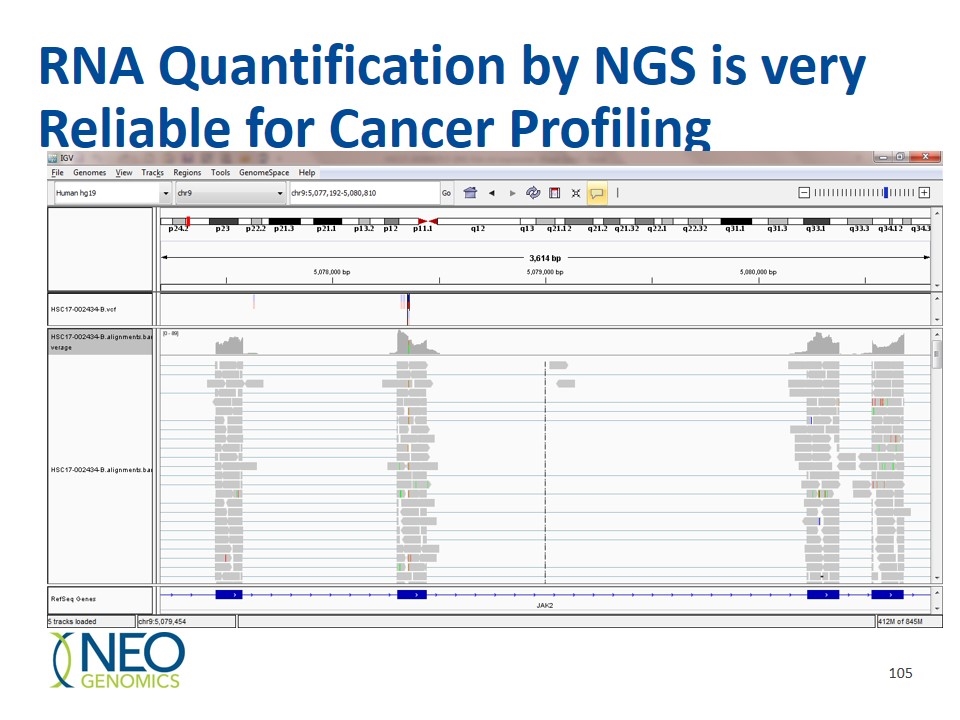
RNA Quantification by NGS is very Reliable for Cancer Profiling
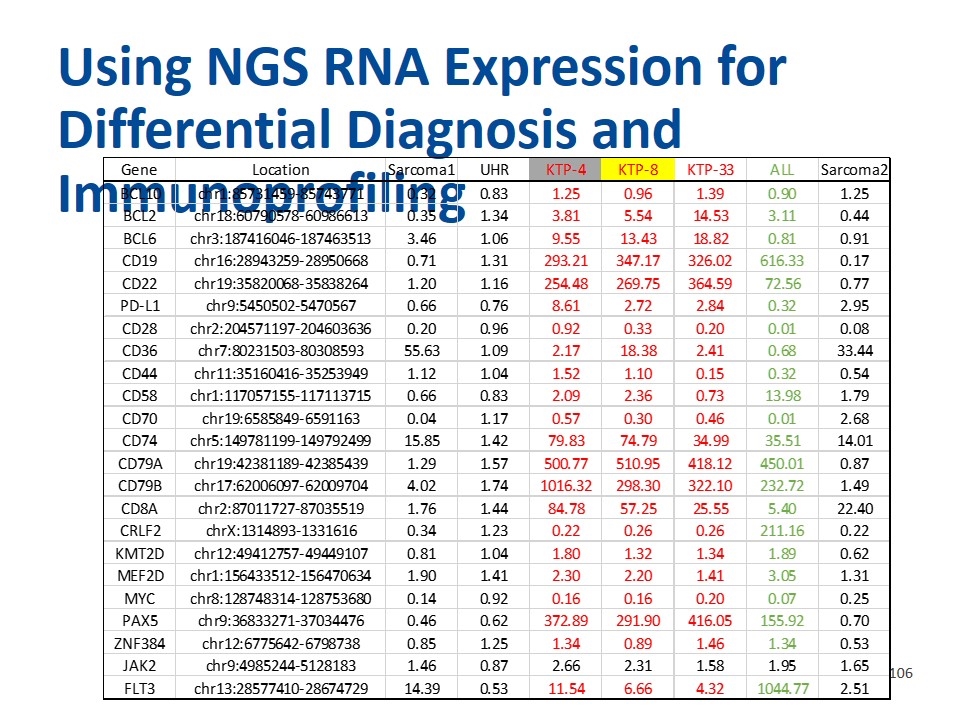
Using NGS RNA Expression for Differential Diagnosis and Immunoprofiling Gene Location Sarcoma1 UHR KTP-4 KTP-8 KTP-33 ALL Sarcoma2 BCL10 chr1:85731459-85743771 0.31673734803505799 0.83298121160716587 1.2485902279795409 0.95631120363944799 1.3917032924666513 0.89958362248438606 1.2472254350116945 BCL2 chr18:60790578-60986613 0.34635290779545591 1.3378696483742827 3.8147607137602724 5.5355041089556316 14.530780386304881 3.1114304630193992 0.43544420857941529 BCL6 chr3:187416046-187463513 3.4640516572067881 1.0578732392328578 9.5526289139544875 13.425882613916391 18.821220348464319 0.81292105301229223 0.90877285199670155 CD19 chr16:28943259-28950668 0.70807685649991337 1.3142721135537476 293.20754716981133 347.16721481737926 326.01956032542842 616.32940972823258 0.16617015752120479 CD22 chr19:35820068-35838264 1.2007184638109305 1.1551550960118167 254.47799113737074 269.74771048744458 364.59438700147712 72.560827178729696 0.7675202363367799 PD-L1 chr9:5450502-5470567 0.66376235875706213 0.76325476694915251 8.6069473870056488 2.7197342867231638 2.8361935028248588 0.32478681144067795 2.9547890183615819 CD28 chr2:204571197-204603636 0.19565622035688079 0.95876004510228563 0.92196946646859845 0.33301558892488325 0.19896658493368893 1.3612227730746515E-2 8.3498290766559888E-2 CD36 chr7:80231503-80308593 55.62721490950512 1.0917526262498418 2.1725335400582204 18.378448930515123 2.4058188836856091 0.67721016327047201 33.439517149727884 CD44 chr11:35160416-35253949 1.1174641067886084 1.0392891967317843 1.5236794324333411 1.1039810362798828 0.14622238660435088 0.32477135940284452 0.53983137755959787 CD58 chr1:117057155-117113715 0.65849196276054067 0.82560465043359244 2.0883598741961737 2.3606349085168423 0.7325959785855386 13.979384422847001 1.7852473610234443 CD70 chr19:6585849-6591163 3.6236762310487412E-2 1.1682345570730486 0.5669414860293196 0.30216865054504449 0.45557398822202733 1.3181105124671093E-2 2.6816839994988095 CD74 chr5:149781199-149792499 15.852055278453012 1.4164767135731651 79.832448754720801 74.79077122357829 34.993186922255219 35.514513537837814 14.014963537417254 CD79A chr19:42381189-42385439 1.2863970385104149 1.5727084369302171 500.77076081551468 510.94811868059008 418.1197856235151 450.01381291784077 0.86887949610475712 CD79B chr17:62006097-62009704 4.0175013963880088 1.7443381120834109 1016.3247067585179 298.29752373859611 322.10426363805618 232.72463228449075 1.4919791472723887 CD8A chr2:87011727-87035519 1.7584216500299286 1.4437972992104275 84.781793389846086 57.254304895646797 25.552439875516303 5.4006850080596491 22.396658909202902 CRLF2 chrX:1314893-1331616 0.33916430887088544 1.2312492807184781 0.21753461779822769 0.26155790406250201 0.26438074475223056 211.16368350310188 0.21587999887086373 KMT2D chr12:49412757-49449107 0.81148119225403403 1.0419324553167968 1.8049908308399654 1.3157788521440041 1.3377760619115726 1.8944537549684564 0.62472514455685313 MEF2D chr1:156433512-156470634 1.8956097640743874 1.4060814926556862 2.3029217819313117 2.2004120828580893 1.4112161758874295 3.0533949822371036 1.3094913965439003 MYC chr8:128748314-128753680 0.14233506223075526 0.92472514351758983 0.16083235426158887 0.15851314821713422 0.20256660075842237 7.0902465350412155E-2 0.25253078106045407 PAX5 chr9:36833271-37034476 0.46265717216981644 0.61975668002056228 372.8925318682368 291.89504575534824 416.04506343281156 155.92368541489984 0.7016109266244287 ZNF384 chr12:6775642-6798738 0.84823540088025773 1.2501202413902628 1.3425745269749081 0.89005671763691652 1.4596578792141206 1.3351603974771995 0.53219383819592547 JAK2 chr9:4985244-5128183 1.4614300877712747 0.86991393384970184 2.6612301619441827 2.3051362607476116 1.5807866591072821 1.9506046797137098 1.6489831064807512 FLT3 chr13:28577410-28674729 14.386470036773776 0.53389892000615313 11.539419340045685 6.6611181898130543 4.3204107517581471 1044.7685167716863 2.5124877009569309 BIVM-ERCC5 chr13:103436630-103528351 63551.536905504268 74642.960349206842 195047.07295928456 2.4778893975573775 0.93050084411930223 0.98339138237843926 1.4969315122663462
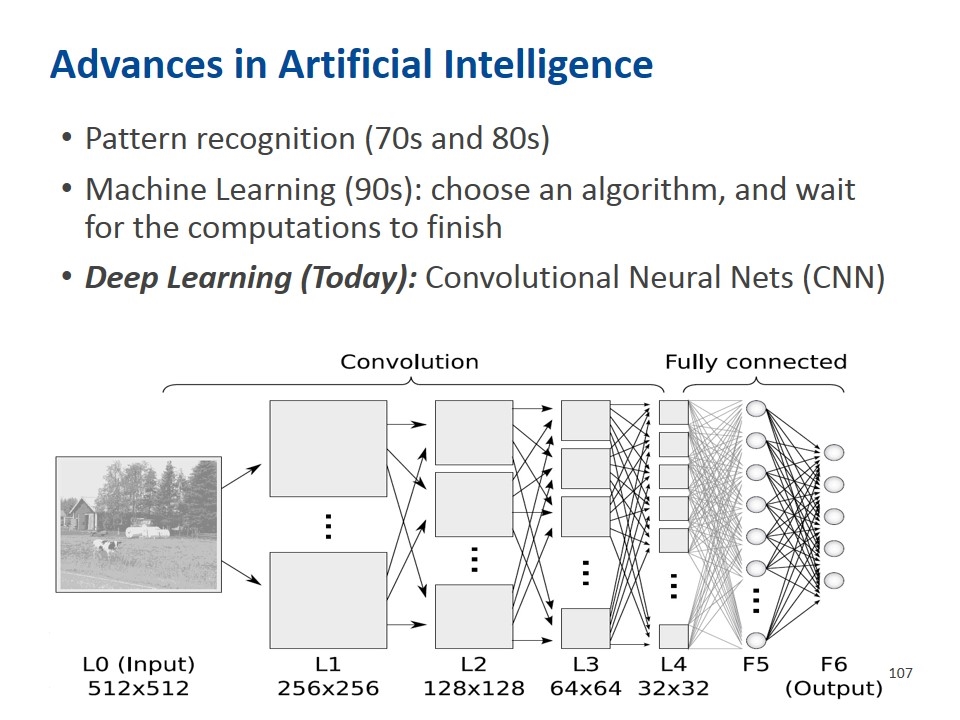
Advances in Artificial Intelligence Pattern recognition (70s and 80s) Machine Learning (90s): choose an algorithm, and wait for the computations to finish Deep Learning (Today): Convolutional Neural Nets (CNN)

Utilizing Deep Learning/Artificial Intelligence (AI) Reducing subjectivity in: Diagnosis Predicting response Predicting prognosis Predicting relapse and MRD Laboratory Processes and procedures
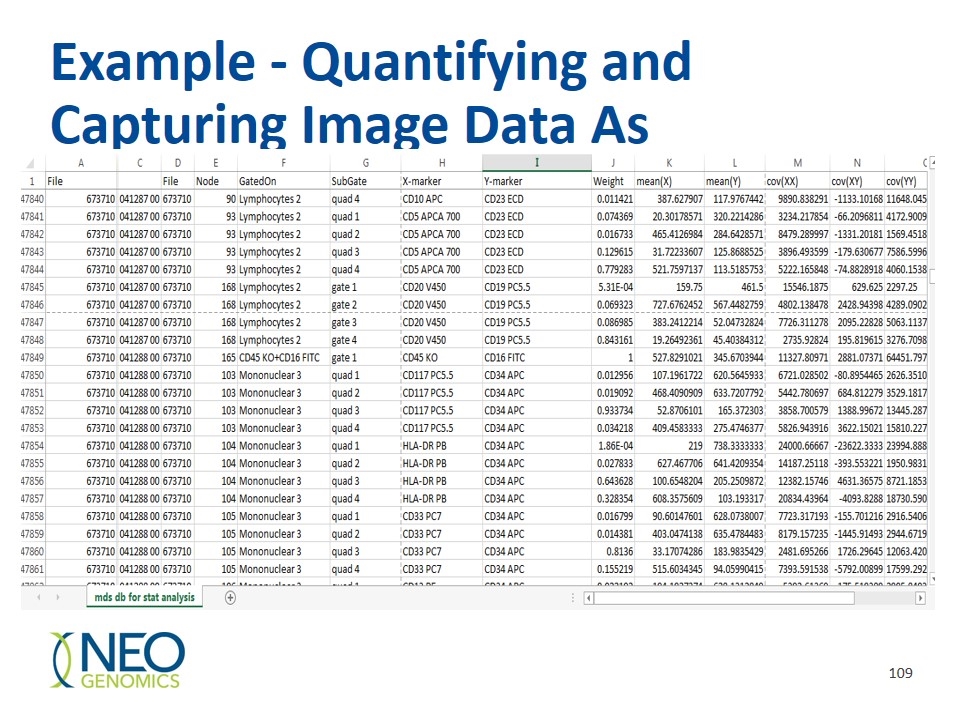
Example - Quantifying and Capturing Image Data As Numerical Values
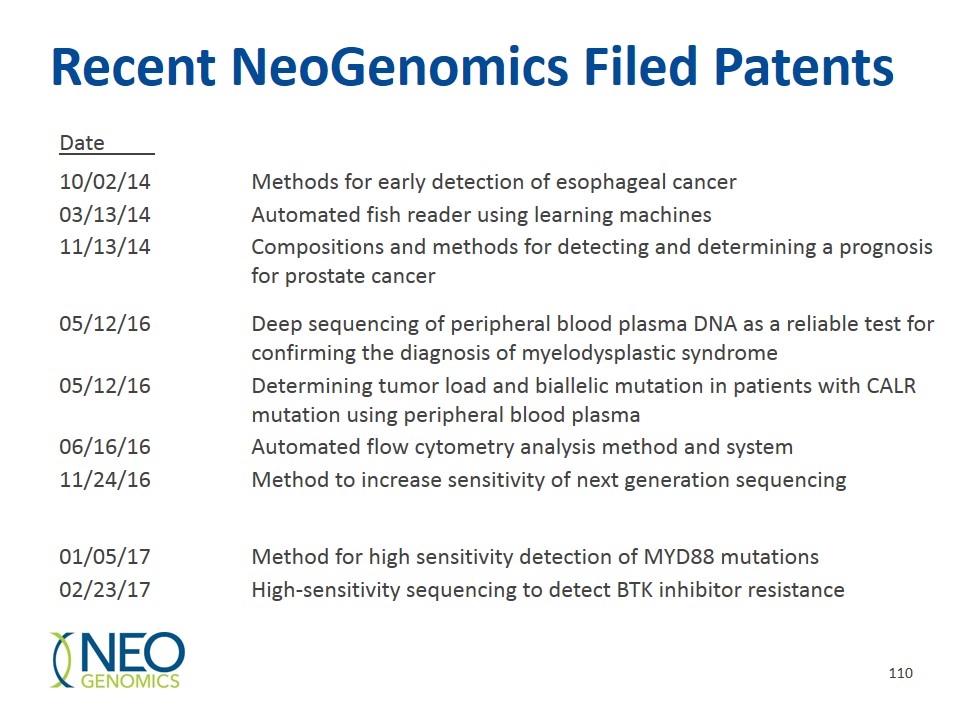
Recent NeoGenomics Filed Patents Date 10/02/14Methods for early detection of esophageal cancer 03/13/14Automated fish reader using learning machines 11/13/14Compositions and methods for detecting and determining a prognosis for prostate cancer 05/12/16Deep sequencing of peripheral blood plasma DNA as a reliable test for confirming the diagnosis of myelodysplastic syndrome 05/12/16Determining tumor load and biallelic mutation in patients with CALR mutation using peripheral blood plasma 06/16/16Automated flow cytometry analysis method and system 11/24/16Method to increase sensitivity of next generation sequencing 01/05/17Method for high sensitivity detection of MYD88 mutations 02/23/17High-sensitivity sequencing to detect BTK inhibitor resistance
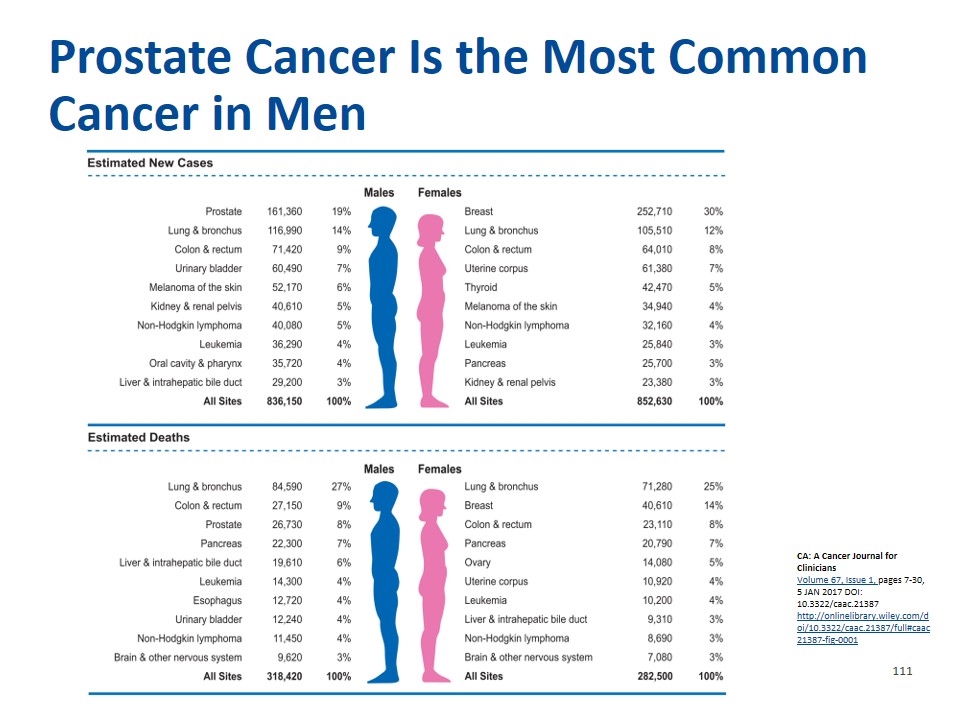
CA: A Cancer Journal for Clinicians Volume 67, Issue 1, pages 7-30, 5 JAN 2017 DOI: 10.3322/caac.21387 http://onlinelibrary.wiley.com/doi/10.3322/caac.21387/full#caac21387-fig-0001 Prostate Cancer Is the Most Common Cancer in Men
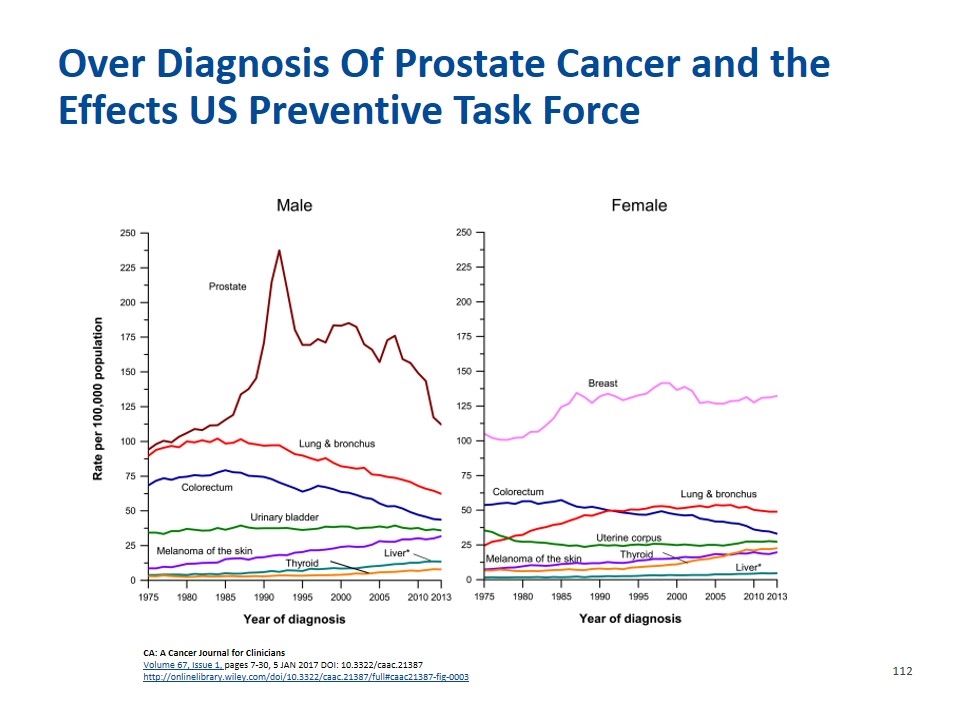
CA: A Cancer Journal for Clinicians Volume 67, Issue 1, pages 7-30, 5 JAN 2017 DOI: 10.3322/caac.21387 http://onlinelibrary.wiley.com/doi/10.3322/caac.21387/full#caac21387-fig-0003 Over Diagnosis Of Prostate Cancer and the Effects US Preventive Task Force

Currently Unmet Needs in Prostate Cancer Screening and Management Better Screening approach Reducing unnecessary biopsies Reducing overtreatment Better selection of patients for active Surveillance better monitoring
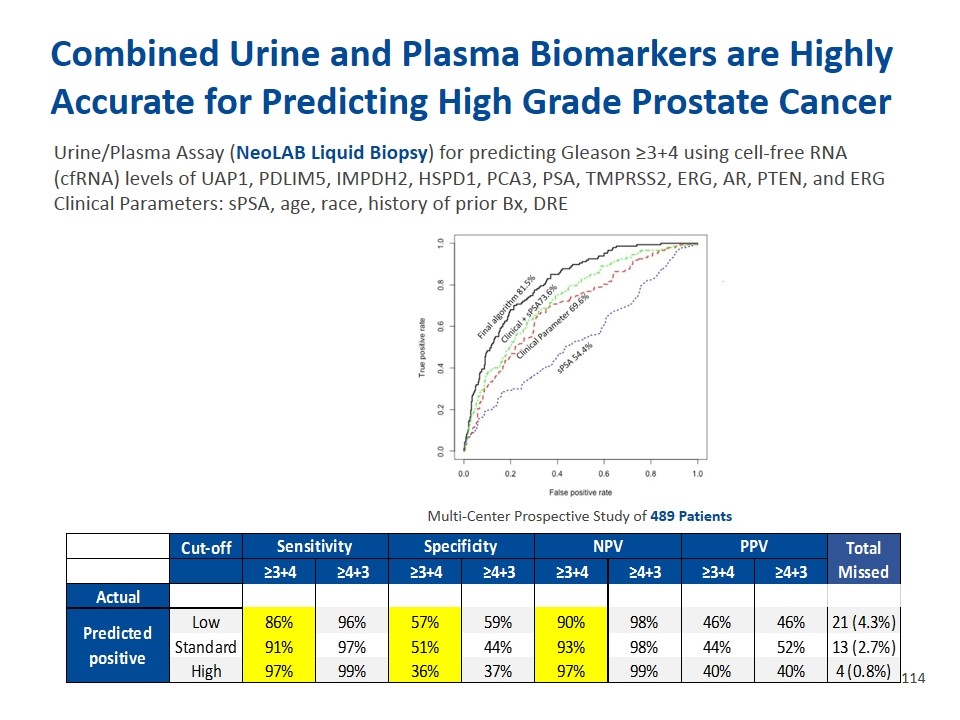
Urine/Plasma Assay (NeoLAB Liquid Biopsy) for predicting Gleason ≥3+4 using cell-free RNA (cfRNA) levels of UAP1, PDLIM5, IMPDH2, HSPD1, PCA3, PSA, TMPRSS2, ERG, AR, PTEN, and ERG Clinical Parameters: sPSA, age, race, history of prior Bx, DRE Multi-Center Prospective Study of 489 Patients Combined Urine and Plasma Biomarkers are Highly Accurate for Predicting High Grade Prostate Cancer Estimated 95% Confidence Interval Value Lower Limit Upper Limit Specificity PPV NPV Prevalence 0.3 0.26 0.34 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Sensitivity 0.97 0.93 0.99 0.99 0.36 0.31 0.42 0.4 0.35 0.45 0.97 0.92 0.99 Specificity 0.36 0.31 0.42 1 0.37 0.32 0.42 0.4 0.35 0.45 0.99 0.95 1 For any particular test result, the probability that it will be: 0.95 0.51 0.45 0.56000000000000005 0.44 0.39 0.5 0.93 0.88 0.96 Positive 0.74 0.7 0.78 0.99 0.44 0.39 0.5 0.52 0.46 0.56999999999999995 0.98 0.94 0.99 Negative 0.26 0.22 0.3 0.91 0.56999999999999995 0.52 0.62 0.46 0.4 0.52 0.9 0.85 0.94 For any particular positive test result, the probability that it is: 0.99 0.59 0.54 0.64 0.46 0.4 0.52 0.98 0.94 0.99 True Positive 0.4 0.35 0.45 False Positive 0.6 0.55000000000000004 0.65 For any particular negative test result, the probability that it is: True Negative 0.97 0.92 0.99 False Negative 0.03 0.01 0.08 likelihood Ratios: [C] = conventional Yes No Yes [W] = weighted by prevalence Neg 106 91 148 Positive [C] 1.53 1.4 1.66 3+3 63 33 70 Negative [C] 0.08 0.03 0.2 3+4 83 3 89 Positive [W] 0.66 0.56000000000000005 0.76 4+3 28 0 29 Negative [W] 0.03 0.01 0.08 >4+3 23 1 25 For 4+3 Cut-off Sensitivity Specificity NPV PPV Total Missed Condition Totals ≥3+4 ≥4+3 ≥3+4 ≥4+3 ≥3+4 ≥4+3 ≥3+4 ≥4+3 Absent Present Actual Test Positive 218 143 361 Predicted positive Low 0.86 0.96 0.56999999999999995 0.59 0.9 0.98 0.46 0.46 21 (4.3%) Test Negative 127 1 128 Standard 0.91 0.97 0.51 0.44 0.93 0.98 0.44 0.52 13 (2.7%) Totals 345 144 489 High 0.97 0.99 0.36 0.37 0.97 0.99 0.4 0.4 4 (0.8%) Estimated 95% Confidence Interval Value Lower Limit Upper Limit Prevalence 0.28999999999999998 0.25 0.34 Sensitivity 0.99 0.96 1 Specificity 0.37 0.32 0.42 For any particular test result, the probability that it will be: Positive 0.74 0.7 0.78 No 146 Negative 0.26 0.22 0.3 For any particular positive test result, the probability that it is: No 133 True Positive 0.4 0.35 0.45 False Positive 0.6 0.55000000000000004 0.65 No 91 For any particular negative test result, the probability that it is: True Negative 0.99 0.95 1 False Negative 0.01 0 0.05 likelihood Ratios: [C] = conventional [W] = weighted by prevalence Positive [C] 1.57 1.45 1.71 Negative [C] 0.02 0 0.13 Positive [W] 0.66 0.56000000000000005 0.76 Negative [W] 0.01 0 0.06 Condition Totals Absent Present Test Positive 169 134 303 Test Negative 173 13 186 Totals 342 147 489 Estimated 95% Confidence Interval Value Lower Limit Upper Limit Prevalence 0.3 0.26 0.34 Sensitivity 0.91 0.85 0.95 Specificity 0.51 0.45 0.56000000000000005 For any particular test result, the probability that it will be: Positive 0.62 0.56999999999999995 0.66 Negative 0.38 0.34 0.43 For any particular positive test result, the probability that it is: True Positive 0.44 0.39 0.5 False Positive 0.56000000000000005 0.5 0.61 For any particular negative test result, the probability that it is: True Negative 0.93 0.88 0.96 False Negative 7.0000000000000007E-2 0.04 0.12 likelihood Ratios: [C] = conventional [W] = weighted by prevalence Positive [C] 1.84 1.64 2.08 Negative [C] 0.17 0.1 0.3 Positive [W] 0.79 0.67 0.93 Negative [W] 0.08 0.04 0.13 For 4+3: Condition Totals Absent Present Test Positive 169 134 303 Test Negative 182 4 186 Totals 351 138 489 Estimated 95% Confidence Interval Value Lower Limit Upper Limit Prevalence 0.28000000000000003 0.24 0.32 Sensitivity 0.97 0.92 0.99 Specificity 0.52 0.46 0.56999999999999995 For any particular test result, the probability that it will be: Positive 0.62 0.56999999999999995 0.66 Negative 0.38 0.34 0.43 For any particular positive test result, the probability that it is: True Positive 0.44 0.39 0.5 False Positive 0.56000000000000005 0.5 0.61 For any particular negative test result, the probability that it is: True Negative 0.98 0.94 0.99 False Negative 0.02 0.01 0.06 likelihood Ratios: [C] = conventional [W] = weighted by prevalence Positive [C] 2.02 1.8 2.2599999999999998 Negative [C] 0.06 0.02 0.15 Positive [W] 0.79 0.67 0.93 Negative [W] 0.02 0.01 0.06 Condition Totals Absent Present Test Positive 147 126 273 Test Negative 195 21 216 Totals 342 147 489 Estimated 95% Confidence Interval Value Lower Limit Upper Limit Prevalence 0.3 0.26 0.34 Sensitivity 0.86 0.79 0.91 Specificity 0.56999999999999995 0.52 0.62 For any particular test result, the probability that it will be: Positive 0.56000000000000005 0.51 0.6 Negative 0.44 0.4 0.49 For any particular positive test result, the probability that it is: True Positive 0.46 0.4 0.52 False Positive 0.54 0.48 0.6 For any particular negative test result, the probability that it is: True Negative 0.9 0.85 0.94 False Negative 0.1 0.06 0.15 likelihood Ratios: [C] = conventional [W] = weighted by prevalence Positive [C] 1.99 1.74 2.29 Negative [C] 0.25 0.17 0.37 Positive [W] 0.86 0.72 1.01 Negative [W] 0.11 7.0000000000000007E-2 0.16 For 4+3 Condition Totals Absent Present Test Positive 147 126 273 Test Negative 211 5 216 Totals 358 131 489 Estimated 95% Confidence Interval Value Lower Limit Upper Limit Prevalence 0.27 0.23 0.31 Sensitivity 0.96 0.91 0.99 Specificity 0.59 0.54 0.64 For any particular test result, the probability that it will be: Positive 0.56000000000000005 0.51 0.6 Negative 0.44 0.4 0.49 For any particular positive test result, the probability that it is: True Positive 0.46 0.4 0.52 False Positive 0.54 0.48 0.6 For any particular negative test result, the probability that it is: True Negative 0.98 0.94 0.99 False Negative 0.02 0.01 0.06 likelihood Ratios: [C] = conventional [W] = weighted by prevalence Positive [C] 2.34 2.06 2.66 Negative [C] 0.06 0.03 0.15 Positive [W] 0.86 0.72 1.01 Negative [W] 0.02 0.01 0.06
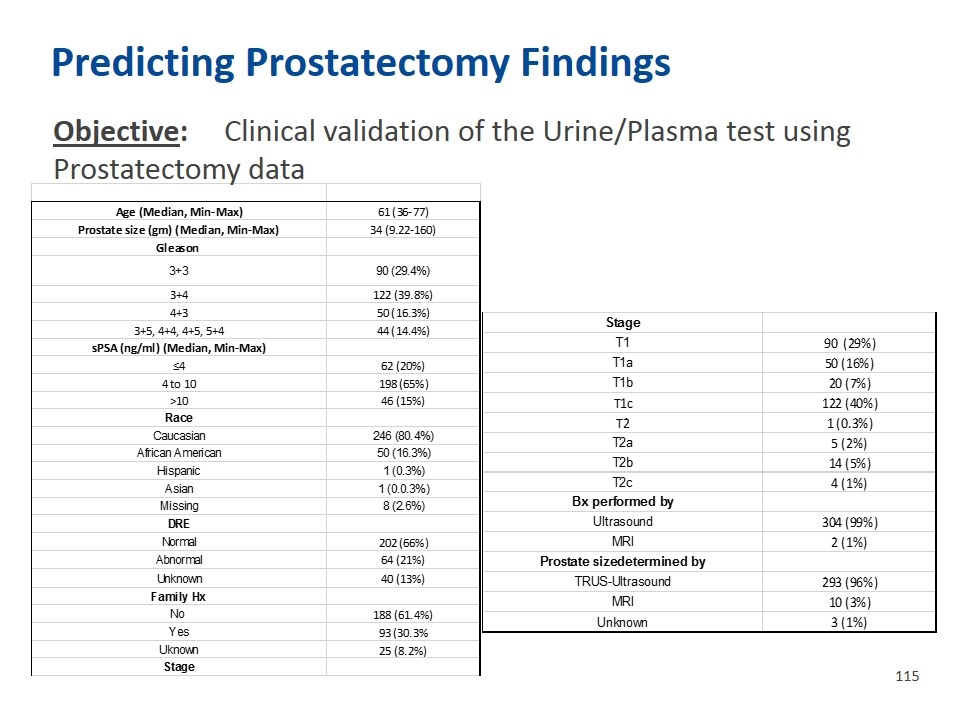
Predicting Prostatectomy Findings Objective: Clinical validation of the Urine/Plasma test using Prostatectomy data Variable Descriptive Statistics (CUSP data,8,16) Valid N Mean Median Minimum Maximum Std.Dev. Age (Median, Min-Max) 61 (36-77) Age (Median, Min-Max) 61 (36-77) Family Hx DRE results 266 1.2406015037593985 1 1 2 0.42825448156307977 Prostate size (gm) (Median, Min-Max) 34 (9.22-160) Prostate size (gm) (Median, Min-Max) 34 (9.22-160) No 188 (61.4%) volume determined TRUS/U-MRI 303 1.0330033003300332 1 1 2 0.17894065571361206 Gleason Gleason Yes 93 (30.3% Prostate Volume CC 291 40.563690343642612 34.299999999999997 9.2200000000000006 160 23.381234482987757 3+3 90 (29.4%) 3+3 90 (29.4%) Uknown 25 (8.2%) Age at Bx 306 60.761437908496745 61 36 77 7.0607631982692647 3+4 122 (39.8%) 3+4 122 (39.8%) Stage No of Cores 306 12.274509803921571 12 4 24 1.8061290701210571 4+3 50 (16.3%) 4+3 50 (16.3%) T1 90 (29%) 3+5, 4+4, 4+5, 5+4 44 (14.4%) 3+5, 4+4, 4+5, 5+4 44 (14.4%) T1a 50 (16%) sPSA (ng/ml) (Median, Min-Max) sPSA (ng/ml) (Median, Min-Max) T1b 20 (7%) ≤4 62 (20%) ≤4 62 (20%) T1c 122 (40%) 4 to 10 198 (65%) 4 to 10 198 (65%) T2 1 (0.3%) >10 46 (15%) >10 46 (15%) T2a 5 (2%) Race Race T2b 14 (5%) Caucasian 246 (80.4%) Caucasian 246 (80.4%) T2c 4 (1%) African American 50 (16.3%) African American 50 (16.3%) Bx performed by Hispanic 1 (0.3%) Hispanic 1 (0.3%) Ultrasound 304 (99%) Asian 1 (0.0.3%) Asian 1 (0.0.3%) MRI 2 (1%) Missing 8 (2.6%) Missing 8 (2.6%) Prostate sizedetermined by DRE DRE TRUS-Ultrasound 293 (96%) Normal 202 (66%) Normal 202 (66%) MRI 10 (3%) Abnormal 64 (21%) Abnormal 64 (21%) Unknown 3 (1%) Unknown 40 (13%) Unknown 40 (13%) Family Hx No 188 (61.4%) Yes 93 (30.3% Uknown 25 (8.2%) Stage T1 90 (29%) T1a 50 (16%) T1b 20 (7%) T1c 122 (40%) T2 1 (0.3%) T2a 5 (2%) T2b 14 (5%) T2c 4 (1%) Bx performed by Ultrasound 304 (99%) MRI 2 (1%) Prostate sizedetermined by TRUS-Ultrasound 293 (96%) MRI 10 (3%) Unknown 3 (1%) Variable Descriptive Statistics (CUSP data,8,16) Valid N Mean Median Minimum Maximum Std.Dev. Age (Median, Min-Max) 61 (36-77) Age (Median, Min-Max) 61 (36-77) Family Hx DRE results 266 1.2406015037593985 1 1 2 0.42825448156307977 Prostate size (gm) (Median, Min-Max) 34 (9.22-160) Prostate size (gm) (Median, Min-Max) 34 (9.22-160) No 188 (61.4%) volume determined TRUS/U-MRI 303 1.0330033003300332 1 1 2 0.17894065571361206 Gleason Gleason Yes 93 (30.3% Prostate Volume CC 291 40.563690343642612 34.299999999999997 9.2200000000000006 160 23.381234482987757 3+3 90 (29.4%) 3+3 90 (29.4%) Uknown 25 (8.2%) Age at Bx 306 60.761437908496745 61 36 77 7.0607631982692647 3+4 122 (39.8%) 3+4 122 (39.8%) Stage No of Cores 306 12.274509803921571 12 4 24 1.8061290701210571 4+3 50 (16.3%) 4+3 50 (16.3%) T1 90 (29%) 3+5, 4+4, 4+5, 5+4 44 (14.4%) 3+5, 4+4, 4+5, 5+4 44 (14.4%) T1a 50 (16%) sPSA (ng/ml) (Median, Min-Max) sPSA (ng/ml) (Median, Min-Max) T1b 20 (7%) ≤4 62 (20%) ≤4 62 (20%) T1c 122 (40%) 4 to 10 198 (65%) 4 to 10 198 (65%) T2 1 (0.3%) >10 46 (15%) >10 46 (15%) T2a 5 (2%) Race Race T2b 14 (5%) Caucasian 246 (80.4%) Caucasian 246 (80.4%) T2c 4 (1%) African American 50 (16.3%) African American 50 (16.3%) Bx performed by Hispanic 1 (0.3%) Hispanic 1 (0.3%) Ultrasound 304 (99%) Asian 1 (0.0.3%) Asian 1 (0.0.3%) MRI 2 (1%) Missing 8 (2.6%) Missing 8 (2.6%) Prostate sizedetermined by DRE DRE TRUS-Ultrasound 293 (96%) Normal 202 (66%) Normal 202 (66%) MRI 10 (3%) Abnormal 64 (21%) Abnormal 64 (21%) Unknown 3 (1%) Unknown 40 (13%) Unknown 40 (13%) Family Hx No 188 (61.4%) Yes 93 (30.3% Uknown 25 (8.2%) Stage T1 90 (29%) T1a 50 (16%) T1b 20 (7%) T1c 122 (40%) T2 1 (0.3%) T2a 5 (2%) T2b 14 (5%) T2c 4 (1%) Bx performed by Ultrasound 304 (99%) MRI 2 (1%) Prostate sizedetermined by TRUS-Ultrasound 293 (96%) MRI 10 (3%) Unknown 3 (1%)
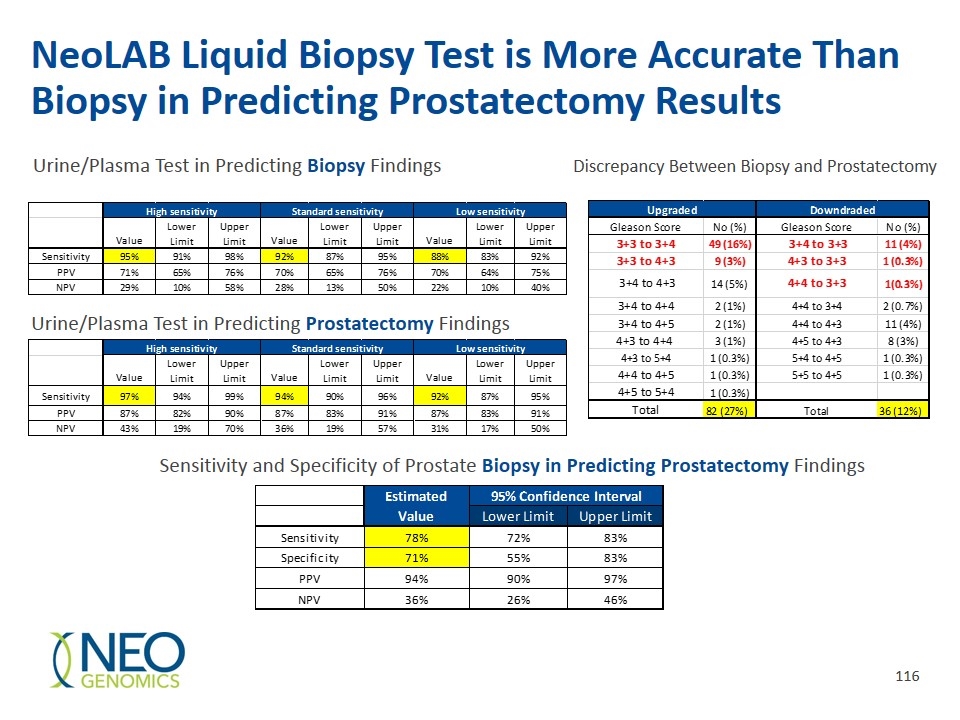
NeoLAB Liquid Biopsy Test is More Accurate Than Biopsy in Predicting Prostatectomy Results Sensitivity and Specificity of Prostate Biopsy in Predicting Prostatectomy Findings Urine/Plasma Test in Predicting Biopsy Findings Urine/Plasma Test in Predicting Prostatectomy Findings Discrepancy Between Biopsy and Prostatectomy Estimated 95% Confidence Interval High sensetivity Standard sensetivity Low sensetivity Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Biopsy Sensitivity 0.92337199999999997 0.88237100000000002 0.95138 0.89814799999999995 0.84794800000000004 0.93365399999999998 0.89655200000000002 0.851495 0.92952000000000001 High Sensitivity Sensitivity 0.92337199999999997 0.88237100000000002 0.95138 Specificity 0.111111 4.1635999999999999E-2 0.24848000000000001 0.111111 5.7472000000000002E-2 0.19917799999999999 0.111111 4.1635999999999999E-2 0.24848000000000001 Specificity 0.111111 4.1635999999999999E-2 0.24848000000000001 Prostatectomy Sensitivity 0.96296300000000001 0.92566700000000002 0.98267700000000002 0.95769199999999999 0.92345200000000005 0.97760199999999997 0.92592600000000003 0.88032600000000005 0.95568500000000001 Standard Sensitivity Sensitivity 0.89814799999999995 0.84794800000000004 0.93365399999999998 Specificity 6.6667000000000004E-2 2.7390999999999999E-2 0.144956 4.3478000000000003E-2 7.5669999999999999E-3 0.16039600000000001 0.1 4.9591999999999997E-2 0.18593299999999999 Specificity 0.111111 5.7472000000000002E-2 0.19917799999999999 Low Sensitivity Sensitivity 0.89655200000000002 0.851495 0.92952000000000001 Specificity 0.111111 4.1635999999999999E-2 0.24848000000000001 Condition Totals Estimated 95% Confidence Interval Absent Present Value Lower Limit Upper Limit Test Positive 13 203 216 Sensitivity 0.96296300000000001 0.92566700000000002 0.98267700000000002 Test Negative 32 58 90 Specificity 6.6667000000000004E-2 2.7390999999999999E-2 0.144956 Totals 45 261 306 Estimated 95% Confidence Interval Sensitivity 0.95769199999999999 0.92345200000000005 0.97760199999999997 Value Lower Limit Upper Limit Specificity 4.3478000000000003E-2 7.5669999999999999E-3 0.16039600000000001 Sensitivity 0.77777799999999997 0.72147799999999995 0.82571600000000001 Specificity 0.71111100000000005 0.55479299999999998 0.83156799999999997 Sensitivity 0.92592600000000003 0.88032600000000005 0.95568500000000001 PPV 0.93981499999999996 0.896957 0.96622200000000003 Specificity 0.1 4.9591999999999997E-2 0.18593299999999999 NPV 0.35555599999999998 0.25944200000000001 0.464144 Bx Estimated 95% Confidence Interval High sensetivity Standard sensetivity Low sensetivity High sensitivity Standard sensitivity Low sensitivity Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Biopsy Sensitivity 0.953704 0.91400000000000003 0.97630300000000003 0.91666700000000001 0.86941800000000002 0.948465 0.88425900000000002 0.832098 0.92227800000000004 Sensitivity 0.953704 0.91400000000000003 0.97630300000000003 0.91666700000000001 0.86941800000000002 0.948465 0.88425900000000002 0.832098 0.92227800000000004 High Sensitivity Sensitivity 0.92337199999999997 0.88237100000000002 0.95138 Specificity 4.4443999999999997E-2 1.4333E-2 0.11620999999999999 7.7778E-2 3.4511E-2 0.15885299999999999 7.7778E-2 3.4511E-2 0.15885299999999999 PPV 0.70547899999999997 0.64902499999999996 0.75642500000000001 0.70462599999999997 0.64696900000000002 0.75658199999999998 0.69708000000000003 0.63836499999999996 0.75016499999999997 Specificity 0.111111 4.1635999999999999E-2 0.24848000000000001 NPV 0.28571400000000002 9.5818E-2 0.57996800000000004 0.28000000000000003 0.12872400000000001 0.49598999999999999 0.21875 9.9441000000000002E-2 0.404418 Prostatectomy Sensitivity 0.96934900000000002 0.938226 0.98567899999999997 0.938697 0.90046099999999996 0.963395 0.915709 0.87346000000000001 0.94522899999999999 Prostatectomy Standard Sensitivity Sensitivity 0.89814799999999995 0.84794800000000004 0.93365399999999998 Specificity 0.13333300000000001 5.5390000000000002E-2 0.27488800000000002 0.2 0.10089099999999999 0.35051599999999999 0.222222 0.117149 0.37476300000000001 High sensetivity Standard sensetivity Low sensetivity Specificity 0.111111 5.7472000000000002E-2 0.19917799999999999 Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 0.96934900000000002 0.938226 0.98567899999999997 0.938697 0.90046099999999996 0.963395 0.915709 0.87346000000000001 0.94522899999999999 Low Sensitivity Sensitivity 0.89655200000000002 0.851495 0.92952000000000001 PPV 0.86643800000000004 0.82072999999999996 0.90221799999999996 0.87188600000000005 0.82575600000000005 0.90753499999999998 0.87226300000000001 0.82549799999999995 0.90826499999999999 Specificity 0.111111 4.1635999999999999E-2 0.24848000000000001 NPV 0.42857099999999998 0.188137 0.70351799999999998 0.36 0.18712000000000001 0.573847 0.3125 0.167495 0.50136199999999997 Condition Totals Estimated 95% Confidence Interval Absent Present Value Lower Limit Upper Limit Test Positive 13 203 216 Sensitivity 0.96296300000000001 0.92566700000000002 0.98267700000000002 Test Negative 32 58 90 Specificity 6.6667000000000004E-2 2.7390999999999999E-2 0.144956 Totals 45 261 306 Estimated 95% Confidence Interval Sensitivity 0.95769199999999999 0.92345200000000005 0.97760199999999997 Value Lower Limit Upper Limit Specificity 4.3478000000000003E-2 7.5669999999999999E-3 0.16039600000000001 Sensitivity 0.77777799999999997 0.72147799999999995 0.82571600000000001 Specificity 0.71111100000000005 0.55479299999999998 0.83156799999999997 Sensitivity 0.92592600000000003 0.88032600000000005 0.95568500000000001 PPV 0.93981499999999996 0.896957 0.96622200000000003 Specificity 0.1 4.9591999999999997E-2 0.18593299999999999 NPV 0.35555599999999998 0.25944200000000001 0.464144 Bx Estimated 95% Confidence Interval High sensetivity Standard sensetivity Low sensetivity High sensetivity Standard sensetivity Low sensetivity Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Biopsy Sensitivity 0.953704 0.91400000000000003 0.97630300000000003 0.91666700000000001 0.86941800000000002 0.948465 0.88425900000000002 0.832098 0.92227800000000004 Sensitivity 0.953704 0.91400000000000003 0.97630300000000003 0.91666700000000001 0.86941800000000002 0.948465 0.88425900000000002 0.832098 0.92227800000000004 High Sensitivity Sensitivity 0.92337199999999997 0.88237100000000002 0.95138 Specificity 4.4443999999999997E-2 1.4333E-2 0.11620999999999999 7.7778E-2 3.4511E-2 0.15885299999999999 7.7778E-2 3.4511E-2 0.15885299999999999 PPV 0.70547899999999997 0.64902499999999996 0.75642500000000001 0.70462599999999997 0.64696900000000002 0.75658199999999998 0.69708000000000003 0.63836499999999996 0.75016499999999997 Specificity 0.111111 4.1635999999999999E-2 0.24848000000000001 NPV 0.28571400000000002 9.5818E-2 0.57996800000000004 0.28000000000000003 0.12872400000000001 0.49598999999999999 0.21875 9.9441000000000002E-2 0.404418 Prostatectomy Sensitivity 0.96934900000000002 0.938226 0.98567899999999997 0.938697 0.90046099999999996 0.963395 0.915709 0.87346000000000001 0.94522899999999999 Prostatectomy Standard Sensitivity Sensitivity 0.89814799999999995 0.84794800000000004 0.93365399999999998 Specificity 0.13333300000000001 5.5390000000000002E-2 0.27488800000000002 0.2 0.10089099999999999 0.35051599999999999 0.222222 0.117149 0.37476300000000001 High sensitivity Standard sensitivity Low sensitivity Specificity 0.111111 5.7472000000000002E-2 0.19917799999999999 Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 0.96934900000000002 0.938226 0.98567899999999997 0.938697 0.90046099999999996 0.963395 0.915709 0.87346000000000001 0.94522899999999999 Low Sensitivity Sensitivity 0.89655200000000002 0.851495 0.92952000000000001 PPV 0.86643800000000004 0.82072999999999996 0.90221799999999996 0.87188600000000005 0.82575600000000005 0.90753499999999998 0.87226300000000001 0.82549799999999995 0.90826499999999999 Specificity 0.111111 4.1635999999999999E-2 0.24848000000000001 NPV 0.42857099999999998 0.188137 0.70351799999999998 0.36 0.18712000000000001 0.573847 0.3125 0.167495 0.50136199999999997 Condition Totals Estimated 95% Confidence Interval Absent Present Value Lower Limit Upper Limit Test Positive 13 203 216 Sensitivity 0.96296300000000001 0.92566700000000002 0.98267700000000002 Test Negative 32 58 90 Specificity 6.6667000000000004E-2 2.7390999999999999E-2 0.144956 Totals 45 261 306 Estimated 95% Confidence Interval Sensitivity 0.95769199999999999 0.92345200000000005 0.97760199999999997 Value Lower Limit Upper Limit Specificity 4.3478000000000003E-2 7.5669999999999999E-3 0.16039600000000001 Sensitivity 0.77777799999999997 0.72147799999999995 0.82571600000000001 Specificity 0.71111100000000005 0.55479299999999998 0.83156799999999997 Sensitivity 0.92592600000000003 0.88032600000000005 0.95568500000000001 PPV 0.93981499999999996 0.896957 0.96622200000000003 Specificity 0.1 4.9591999999999997E-2 0.18593299999999999 NPV 0.35555599999999998 0.25944200000000001 0.464144 Variable Descriptive Statistics (CUSP data,8,16) Upgraded Downdraded Valid N Mean Median Minimum Maximum Std.Dev. Age (Median, Min-Max) 61 (36-77) Age (Median, Min-Max) 61 (36-77) Family Hx Gleason Score No (%) Gleason Score No (%) DRE results 266 1.2406015037593985 1 1 2 0.42825448156307977 Prostate size (gm) (Median, Min-Max) 34 (9.22-160) Prostate size (gm) (Median, Min-Max) 34 (9.22-160) No 188 (61.4%) 3+3 to 3+4 49 (16%) 3+4 to 3+3 11 (4%) volume determined TRUS/U-MRI 303 1.0330033003300332 1 1 2 0.17894065571361206 Gleason Score by Biopsy Gleason Yes 93 (30.3% 3+3 to 4+3 9 (3%) 4+3 to 3+3 1 (0.3%) Prostate Volume CC 291 40.563690343642612 34.299999999999997 9.2200000000000006 160 23.381234482987757 3+3 90 (29.4%) 3+3 90 (29.4%) Uknown 25 (8.2%) 3+4 to 4+3 14 (5%) 4+4 to 3+3 1(0.3%) Age at Bx 306 60.761437908496745 61 36 77 7.0607631982692647 3+4 122 (39.8%) 3+4 122 (39.8%) Stage 3+4 to 4+4 2 (1%) 4+4 to 3+4 2 (0.7%) No of Cores 306 12.274509803921571 12 4 24 1.8061290701210571 4+3 50 (16.3%) 4+3 50 (16.3%) T1 90 (29%) 3+4 to 4+5 2 (1%) 4+4 to 4+3 11 (4%) 3+5, 4+4, 4+5, 5+4 44 (14.4%) 3+5, 4+4, 4+5, 5+4 44 (14.4%) T1a 50 (16%) 4+3 to 4+4 3 (1%) 4+5 to 4+3 8 (3%) sPSA (ng/ml) (Median, Min-Max) 5.51 (0.58-46.0) sPSA (ng/ml) (Median, Min-Max) T1b 20 (7%) 4+3 to 5+4 1 (0.3%) 5+4 to 4+5 1 (0.3%) Variable Descriptive Statistics (CUSP data,8,16) ≤4 62 (20%) ≤4 62 (20%) T1c 122 (40%) 4+4 to 4+5 1 (0.3%) 5+5 to 4+5 1 (0.3%) Valid N Mean Median Minimum Maximum Std.Dev. 4 to 10 198 (65%) 4 to 10 198 (65%) T2 1 (0.3%) 4+5 to 5+4 1 (0.3%) PSA 306 6.9517745098039221 5.5049999999999999 0.57999999999999996 46 5.2179599969063899 >10 46 (15%) >10 46 (15%) T2a 5 (2%) Total 82 (27%) Total 36 (12%) Race Race T2b 14 (5%) Caucasian 246 (80.4%) Caucasian 246 (80.4%) T2c 4 (1%) African American 50 (16.3%) African American 50 (16.3%) Bx performed by Hispanic 1 (0.3%) Hispanic 1 (0.3%) Ultrasound 304 (99%) Asian 1 (0.0.3%) Asian 1 (0.0.3%) MRI 2 (1%) Missing 8 (2.6%) Missing 8 (2.6%) Prostate sizedetermined by DRE DRE TRUS-Ultrasound 293 (96%) Normal 202 (66%) Normal 202 (66%) MRI 10 (3%) Abnormal 64 (21%) Abnormal 64 (21%) Unknown 3 (1%) Unknown 40 (13%) Unknown 40 (13%) Family Hx No 188 (61.4%) Yes 93 (30.3% Uknown 25 (8.2%) Stage T1 90 (29%) T1a 50 (16%) T1b 20 (7%) T1c 122 (40%) T2 1 (0.3%) T2a 5 (2%) T2b 14 (5%) T2c 4 (1%) Bx performed by Ultrasound 304 (99%) MRI 2 (1%) Prostate size was determined by TRUS-Ultrasound 293 (96%) MRI 10 (3%) Unknown 3 (1%)

NeoLAB Prostate Test Indications Prior to performing biopsy When Biopsy findings are questionable To determine if patients patient should be put on active surveillance To monitor patients on Active Surveillance
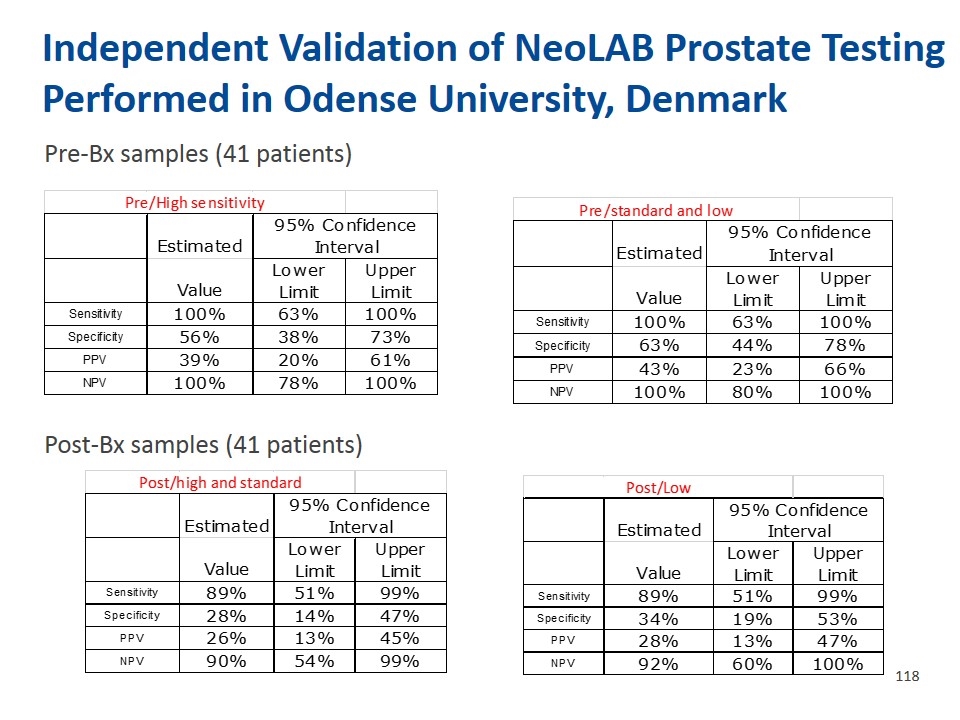
Pre-Bx samples (41 patients) Post-Bx samples (41 patients) Independent Validation of NeoLAB Prostate Testing Performed in Odense University, Denmark High Standard Low Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Test Positive 42 12 54 Test Positive 40 12 52 Test Positive 38 12 50 Test Positive 14 9 23 Test Positive 12 9 21 Test Positive 23 8 31 Test Positive 21 8 29 Test Negative 27 1 28 Test Negative 29 1 30 Test Negative 31 1 32 Test Negative 18 0 18 Test Negative 20 0 20 Test Negative 9 1 10 Test Negative 11 1 12 Totals 69 13 82 Totals 69 13 82 Totals 69 13 82 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: Positive 0.65853700000000004 0.544651 0.75741099999999995 Positive 0.63414599999999999 0.51991399999999999 0.73568699999999998 Positive 0.60975599999999996 0.49544199999999999 0.71369300000000002 Positive 0.56097600000000003 0.398895 0.71180699999999997 Positive 0.51219499999999996 0.353655 0.66848600000000002 Positive 0.75609800000000005 0.59355800000000003 0.87092199999999997 Positive 0.70731699999999997 0.542632 0.83353200000000005 Negative 0.34146300000000002 0.242589 0.455349 Negative 0.36585400000000001 0.26431300000000002 0.48008600000000001 Negative 0.39024399999999998 0.28630699999999998 0.50455799999999995 Negative 0.43902400000000003 0.28819299999999998 0.601105 Negative 0.48780499999999999 0.33151399999999998 0.64634499999999995 Negative 0.24390200000000001 0.129078 0.40644200000000003 Negative 0.29268300000000003 0.166468 0.457368 For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: True Positive 0.222222 0.12481200000000001 0.35946400000000001 True Positive 0.230769 0.129806 0.37173600000000001 True Positive 0.24 0.135217 0.38486599999999999 True Positive 0.39130399999999999 0.204677 0.61217500000000002 True Positive 0.42857099999999998 0.225908 0.65561000000000003 True Positive 0.25806499999999999 0.12536900000000001 0.44929599999999997 True Positive 0.275862 0.134459 0.47485100000000002 False Positive 0.77777799999999997 0.64053599999999999 0.87518799999999997 False Positive 0.769231 0.62826400000000004 0.87019400000000002 False Positive 0.76 0.61513399999999996 0.86478299999999997 False Positive 0.60869600000000001 0.38782499999999998 0.795323 False Positive 0.57142899999999996 0.34438999999999997 0.774092 False Positive 0.74193500000000001 0.55070399999999997 0.87463100000000005 False Positive 0.72413799999999995 0.52514899999999998 0.865541 For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: True Negative 0.96428599999999998 0.79761000000000004 0.99813300000000005 True Negative 0.96666700000000005 0.80947000000000002 0.99825799999999998 True Negative 0.96875 0.82001900000000005 0.998367 True Negative 1 0.78124400000000005 1 True Negative 1 0.79954700000000001 1 True Negative 0.9 0.54115500000000005 0.99475800000000003 True Negative 0.91666700000000001 0.59753900000000004 0.99563500000000005 False Negative 3.5714000000000003E-2 1.867E-3 0.20238999999999999 False Negative 3.3333000000000002E-2 1.7420000000000001E-3 0.19053 False Negative 3.125E-2 1.6329999999999999E-3 0.179981 False Negative 0 0 0.21875600000000001 False Negative 0 0 0.20045299999999999 False Negative 0.1 5.2420000000000001E-3 0.458845 False Negative 8.3333000000000004E-2 4.365E-3 0.40246100000000001 likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence Positive [C] 1.5923080000000001 1.2339960000000001 2.0546609999999998 Positive [C] 1.676113 1.286368 2.1839439999999999 Positive [C] 2.285714 1.5431299999999999 3.3856449999999998 Positive [C] 2.6666669999999999 1.7049430000000001 4.1708790000000002 Positive [C] 1.236715 0.90096699999999996 1.6975800000000001 Positive [C] 1.3544970000000001 0.96319399999999999 1.904771 Negative [C] 0.18302399999999999 2.6772000000000001E-2 1.2512080000000001 Negative [C] 0.17121600000000001 2.5155E-2 1.165392 Negative [C] 0 0 NaN Negative [C] 0 0 NaN Negative [C] 0.39506200000000002 5.3231000000000001E-2 2.9319839999999999 Negative [C] 0.32323200000000002 4.5311999999999998E-2 2.3057669999999999 Positive [W] 0.3 0.178699 0.50364100000000001 Positive [W] 0.31578899999999999 0.18825900000000001 0.52971100000000004 Positive [W] 0.64285700000000001 0.35071400000000003 1.1783539999999999 Positive [W] 0.75 0.40453699999999998 1.3904780000000001 Positive [W] 0.34782600000000002 0.184888 0.65435900000000002 Positive [W] 0.38095200000000001 0.20268600000000001 0.71600600000000003 Negative [W] 3.4483E-2 5.0090000000000004E-3 0.237396 Negative [W] 3.2258000000000002E-2 4.6769999999999997E-3 0.22248000000000001 Negative [W] 0 0 NaN Negative [W] 0 0 NaN Negative [W] 0.111111 1.6934999999999999E-2 0.72901099999999996 Negative [W] 9.0909000000000004E-2 1.3715E-2 0.60258400000000001 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 PPV 0.222222 0.12481200000000001 0.35946400000000001 PPV 0.230769 0.129806 0.37173600000000001 PPV 0.24 0.135217 0.38486599999999999 NPV 0.96428599999999998 0.79761000000000004 0.99813300000000005 NPV 0.96666700000000005 0.80947000000000002 0.99825799999999998 NPV 0.96875 0.82001900000000005 0.998367 Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 PPV 0.39130399999999999 0.204677 0.61217500000000002 PPV 0.42857099999999998 0.225908 0.65561000000000003 PPV 0.25806499999999999 0.12536900000000001 0.44929599999999997 PPV 0.275862 0.134459 0.47485100000000002 NPV 1 0.78124400000000005 1 NPV 1 0.79954700000000001 1 NPV 0.9 0.54115500000000005 0.99475800000000003 NPV 0.91666700000000001 0.59753900000000004 0.99563500000000005 High Standard Low Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Test Positive 42 12 54 Test Positive 40 12 52 Test Positive 38 12 50 Test Positive 14 9 23 Test Positive 12 9 21 Test Positive 23 8 31 Test Positive 21 8 29 Test Negative 27 1 28 Test Negative 29 1 30 Test Negative 31 1 32 Test Negative 18 0 18 Test Negative 20 0 20 Test Negative 9 1 10 Test Negative 11 1 12 Totals 69 13 82 Totals 69 13 82 Totals 69 13 82 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: Positive 0.65853700000000004 0.544651 0.75741099999999995 Positive 0.63414599999999999 0.51991399999999999 0.73568699999999998 Positive 0.60975599999999996 0.49544199999999999 0.71369300000000002 Positive 0.56097600000000003 0.398895 0.71180699999999997 Positive 0.51219499999999996 0.353655 0.66848600000000002 Positive 0.75609800000000005 0.59355800000000003 0.87092199999999997 Positive 0.70731699999999997 0.542632 0.83353200000000005 Negative 0.34146300000000002 0.242589 0.455349 Negative 0.36585400000000001 0.26431300000000002 0.48008600000000001 Negative 0.39024399999999998 0.28630699999999998 0.50455799999999995 Negative 0.43902400000000003 0.28819299999999998 0.601105 Negative 0.48780499999999999 0.33151399999999998 0.64634499999999995 Negative 0.24390200000000001 0.129078 0.40644200000000003 Negative 0.29268300000000003 0.166468 0.457368 For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: True Positive 0.222222 0.12481200000000001 0.35946400000000001 True Positive 0.230769 0.129806 0.37173600000000001 True Positive 0.24 0.135217 0.38486599999999999 True Positive 0.39130399999999999 0.204677 0.61217500000000002 True Positive 0.42857099999999998 0.225908 0.65561000000000003 True Positive 0.25806499999999999 0.12536900000000001 0.44929599999999997 True Positive 0.275862 0.134459 0.47485100000000002 False Positive 0.77777799999999997 0.64053599999999999 0.87518799999999997 False Positive 0.769231 0.62826400000000004 0.87019400000000002 False Positive 0.76 0.61513399999999996 0.86478299999999997 False Positive 0.60869600000000001 0.38782499999999998 0.795323 False Positive 0.57142899999999996 0.34438999999999997 0.774092 False Positive 0.74193500000000001 0.55070399999999997 0.87463100000000005 False Positive 0.72413799999999995 0.52514899999999998 0.865541 For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: True Negative 0.96428599999999998 0.79761000000000004 0.99813300000000005 True Negative 0.96666700000000005 0.80947000000000002 0.99825799999999998 True Negative 0.96875 0.82001900000000005 0.998367 True Negative 1 0.78124400000000005 1 True Negative 1 0.79954700000000001 1 True Negative 0.9 0.54115500000000005 0.99475800000000003 True Negative 0.91666700000000001 0.59753900000000004 0.99563500000000005 False Negative 3.5714000000000003E-2 1.867E-3 0.20238999999999999 False Negative 3.3333000000000002E-2 1.7420000000000001E-3 0.19053 False Negative 3.125E-2 1.6329999999999999E-3 0.179981 False Negative 0 0 0.21875600000000001 False Negative 0 0 0.20045299999999999 False Negative 0.1 5.2420000000000001E-3 0.458845 False Negative 8.3333000000000004E-2 4.365E-3 0.40246100000000001 likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence Positive [C] 1.5923080000000001 1.2339960000000001 2.0546609999999998 Positive [C] 1.676113 1.286368 2.1839439999999999 Positive [C] 2.285714 1.5431299999999999 3.3856449999999998 Positive [C] 2.6666669999999999 1.7049430000000001 4.1708790000000002 Positive [C] 1.236715 0.90096699999999996 1.6975800000000001 Positive [C] 1.3544970000000001 0.96319399999999999 1.904771 Negative [C] 0.18302399999999999 2.6772000000000001E-2 1.2512080000000001 Negative [C] 0.17121600000000001 2.5155E-2 1.165392 Negative [C] 0 0 NaN Negative [C] 0 0 NaN Negative [C] 0.39506200000000002 5.3231000000000001E-2 2.9319839999999999 Negative [C] 0.32323200000000002 4.5311999999999998E-2 2.3057669999999999 Positive [W] 0.3 0.178699 0.50364100000000001 Positive [W] 0.31578899999999999 0.18825900000000001 0.52971100000000004 Positive [W] 0.64285700000000001 0.35071400000000003 1.1783539999999999 Positive [W] 0.75 0.40453699999999998 1.3904780000000001 Positive [W] 0.34782600000000002 0.184888 0.65435900000000002 Positive [W] 0.38095200000000001 0.20268600000000001 0.71600600000000003 Negative [W] 3.4483E-2 5.0090000000000004E-3 0.237396 Negative [W] 3.2258000000000002E-2 4.6769999999999997E-3 0.22248000000000001 Negative [W] 0 0 NaN Negative [W] 0 0 NaN Negative [W] 0.111111 1.6934999999999999E-2 0.72901099999999996 Negative [W] 9.0909000000000004E-2 1.3715E-2 0.60258400000000001 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 PPV 0.222222 0.12481200000000001 0.35946400000000001 PPV 0.230769 0.129806 0.37173600000000001 PPV 0.24 0.135217 0.38486599999999999 NPV 0.96428599999999998 0.79761000000000004 0.99813300000000005 NPV 0.96666700000000005 0.80947000000000002 0.99825799999999998 NPV 0.96875 0.82001900000000005 0.998367 Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 PPV 0.39130399999999999 0.204677 0.61217500000000002 PPV 0.42857099999999998 0.225908 0.65561000000000003 PPV 0.25806499999999999 0.12536900000000001 0.44929599999999997 PPV 0.275862 0.134459 0.47485100000000002 NPV 1 0.78124400000000005 1 NPV 1 0.79954700000000001 1 NPV 0.9 0.54115500000000005 0.99475800000000003 NPV 0.91666700000000001 0.59753900000000004 0.99563500000000005 High Standard Low Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Test Positive 42 12 54 Test Positive 40 12 52 Test Positive 38 12 50 Test Positive 14 9 23 Test Positive 12 9 21 Test Positive 23 8 31 Test Positive 21 8 29 Test Negative 27 1 28 Test Negative 29 1 30 Test Negative 31 1 32 Test Negative 18 0 18 Test Negative 20 0 20 Test Negative 9 1 10 Test Negative 11 1 12 Totals 69 13 82 Totals 69 13 82 Totals 69 13 82 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: Positive 0.65853700000000004 0.544651 0.75741099999999995 Positive 0.63414599999999999 0.51991399999999999 0.73568699999999998 Positive 0.60975599999999996 0.49544199999999999 0.71369300000000002 Positive 0.56097600000000003 0.398895 0.71180699999999997 Positive 0.51219499999999996 0.353655 0.66848600000000002 Positive 0.75609800000000005 0.59355800000000003 0.87092199999999997 Positive 0.70731699999999997 0.542632 0.83353200000000005 Negative 0.34146300000000002 0.242589 0.455349 Negative 0.36585400000000001 0.26431300000000002 0.48008600000000001 Negative 0.39024399999999998 0.28630699999999998 0.50455799999999995 Negative 0.43902400000000003 0.28819299999999998 0.601105 Negative 0.48780499999999999 0.33151399999999998 0.64634499999999995 Negative 0.24390200000000001 0.129078 0.40644200000000003 Negative 0.29268300000000003 0.166468 0.457368 For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: True Positive 0.222222 0.12481200000000001 0.35946400000000001 True Positive 0.230769 0.129806 0.37173600000000001 True Positive 0.24 0.135217 0.38486599999999999 True Positive 0.39130399999999999 0.204677 0.61217500000000002 True Positive 0.42857099999999998 0.225908 0.65561000000000003 True Positive 0.25806499999999999 0.12536900000000001 0.44929599999999997 True Positive 0.275862 0.134459 0.47485100000000002 False Positive 0.77777799999999997 0.64053599999999999 0.87518799999999997 False Positive 0.769231 0.62826400000000004 0.87019400000000002 False Positive 0.76 0.61513399999999996 0.86478299999999997 False Positive 0.60869600000000001 0.38782499999999998 0.795323 False Positive 0.57142899999999996 0.34438999999999997 0.774092 False Positive 0.74193500000000001 0.55070399999999997 0.87463100000000005 False Positive 0.72413799999999995 0.52514899999999998 0.865541 For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: True Negative 0.96428599999999998 0.79761000000000004 0.99813300000000005 True Negative 0.96666700000000005 0.80947000000000002 0.99825799999999998 True Negative 0.96875 0.82001900000000005 0.998367 True Negative 1 0.78124400000000005 1 True Negative 1 0.79954700000000001 1 True Negative 0.9 0.54115500000000005 0.99475800000000003 True Negative 0.91666700000000001 0.59753900000000004 0.99563500000000005 False Negative 3.5714000000000003E-2 1.867E-3 0.20238999999999999 False Negative 3.3333000000000002E-2 1.7420000000000001E-3 0.19053 False Negative 3.125E-2 1.6329999999999999E-3 0.179981 False Negative 0 0 0.21875600000000001 False Negative 0 0 0.20045299999999999 False Negative 0.1 5.2420000000000001E-3 0.458845 False Negative 8.3333000000000004E-2 4.365E-3 0.40246100000000001 likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence Positive [C] 1.5923080000000001 1.2339960000000001 2.0546609999999998 Positive [C] 1.676113 1.286368 2.1839439999999999 Positive [C] 2.285714 1.5431299999999999 3.3856449999999998 Positive [C] 2.6666669999999999 1.7049430000000001 4.1708790000000002 Positive [C] 1.236715 0.90096699999999996 1.6975800000000001 Positive [C] 1.3544970000000001 0.96319399999999999 1.904771 Negative [C] 0.18302399999999999 2.6772000000000001E-2 1.2512080000000001 Negative [C] 0.17121600000000001 2.5155E-2 1.165392 Negative [C] 0 0 NaN Negative [C] 0 0 NaN Negative [C] 0.39506200000000002 5.3231000000000001E-2 2.9319839999999999 Negative [C] 0.32323200000000002 4.5311999999999998E-2 2.3057669999999999 Positive [W] 0.3 0.178699 0.50364100000000001 Positive [W] 0.31578899999999999 0.18825900000000001 0.52971100000000004 Positive [W] 0.64285700000000001 0.35071400000000003 1.1783539999999999 Positive [W] 0.75 0.40453699999999998 1.3904780000000001 Positive [W] 0.34782600000000002 0.184888 0.65435900000000002 Positive [W] 0.38095200000000001 0.20268600000000001 0.71600600000000003 Negative [W] 3.4483E-2 5.0090000000000004E-3 0.237396 Negative [W] 3.2258000000000002E-2 4.6769999999999997E-3 0.22248000000000001 Negative [W] 0 0 NaN Negative [W] 0 0 NaN Negative [W] 0.111111 1.6934999999999999E-2 0.72901099999999996 Negative [W] 9.0909000000000004E-2 1.3715E-2 0.60258400000000001 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 PPV 0.222222 0.12481200000000001 0.35946400000000001 PPV 0.230769 0.129806 0.37173600000000001 PPV 0.24 0.135217 0.38486599999999999 NPV 0.96428599999999998 0.79761000000000004 0.99813300000000005 NPV 0.96666700000000005 0.80947000000000002 0.99825799999999998 NPV 0.96875 0.82001900000000005 0.998367 Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 PPV 0.39130399999999999 0.204677 0.61217500000000002 PPV 0.42857099999999998 0.225908 0.65561000000000003 PPV 0.25806499999999999 0.12536900000000001 0.44929599999999997 PPV 0.275862 0.134459 0.47485100000000002 NPV 1 0.78124400000000005 1 NPV 1 0.79954700000000001 1 NPV 0.9 0.54115500000000005 0.99475800000000003 NPV 0.91666700000000001 0.59753900000000004 0.99563500000000005 High Standard Low Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Condition Totals Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Absent Present Test Positive 42 12 54 Test Positive 40 12 52 Test Positive 38 12 50 Test Positive 14 9 23 Test Positive 12 9 21 Test Positive 23 8 31 Test Positive 21 8 29 Test Negative 27 1 28 Test Negative 29 1 30 Test Negative 31 1 32 Test Negative 18 0 18 Test Negative 20 0 20 Test Negative 9 1 10 Test Negative 11 1 12 Totals 69 13 82 Totals 69 13 82 Totals 69 13 82 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Totals 32 9 41 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.15853700000000001 9.0380000000000002E-2 0.259515 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Prevalence 0.21951200000000001 0.11111799999999999 0.38030399999999998 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: For any particular test result, the probability that it will be: Positive 0.65853700000000004 0.544651 0.75741099999999995 Positive 0.63414599999999999 0.51991399999999999 0.73568699999999998 Positive 0.60975599999999996 0.49544199999999999 0.71369300000000002 Positive 0.56097600000000003 0.398895 0.71180699999999997 Positive 0.51219499999999996 0.353655 0.66848600000000002 Positive 0.75609800000000005 0.59355800000000003 0.87092199999999997 Positive 0.70731699999999997 0.542632 0.83353200000000005 Negative 0.34146300000000002 0.242589 0.455349 Negative 0.36585400000000001 0.26431300000000002 0.48008600000000001 Negative 0.39024399999999998 0.28630699999999998 0.50455799999999995 Negative 0.43902400000000003 0.28819299999999998 0.601105 Negative 0.48780499999999999 0.33151399999999998 0.64634499999999995 Negative 0.24390200000000001 0.129078 0.40644200000000003 Negative 0.29268300000000003 0.166468 0.457368 For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: For any particular positive test result, the probability that it is: True Positive 0.222222 0.12481200000000001 0.35946400000000001 True Positive 0.230769 0.129806 0.37173600000000001 True Positive 0.24 0.135217 0.38486599999999999 True Positive 0.39130399999999999 0.204677 0.61217500000000002 True Positive 0.42857099999999998 0.225908 0.65561000000000003 True Positive 0.25806499999999999 0.12536900000000001 0.44929599999999997 True Positive 0.275862 0.134459 0.47485100000000002 False Positive 0.77777799999999997 0.64053599999999999 0.87518799999999997 False Positive 0.769231 0.62826400000000004 0.87019400000000002 False Positive 0.76 0.61513399999999996 0.86478299999999997 False Positive 0.60869600000000001 0.38782499999999998 0.795323 False Positive 0.57142899999999996 0.34438999999999997 0.774092 False Positive 0.74193500000000001 0.55070399999999997 0.87463100000000005 False Positive 0.72413799999999995 0.52514899999999998 0.865541 For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: For any particular negative test result, the probability that it is: True Negative 0.96428599999999998 0.79761000000000004 0.99813300000000005 True Negative 0.96666700000000005 0.80947000000000002 0.99825799999999998 True Negative 0.96875 0.82001900000000005 0.998367 True Negative 1 0.78124400000000005 1 True Negative 1 0.79954700000000001 1 True Negative 0.9 0.54115500000000005 0.99475800000000003 True Negative 0.91666700000000001 0.59753900000000004 0.99563500000000005 False Negative 3.5714000000000003E-2 1.867E-3 0.20238999999999999 False Negative 3.3333000000000002E-2 1.7420000000000001E-3 0.19053 False Negative 3.125E-2 1.6329999999999999E-3 0.179981 False Negative 0 0 0.21875600000000001 False Negative 0 0 0.20045299999999999 False Negative 0.1 5.2420000000000001E-3 0.458845 False Negative 8.3333000000000004E-2 4.365E-3 0.40246100000000001 likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: likelihood Ratios: [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [C] = conventional [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence [W] = weighted by prevalence Positive [C] 1.5923080000000001 1.2339960000000001 2.0546609999999998 Positive [C] 1.676113 1.286368 2.1839439999999999 Positive [C] 2.285714 1.5431299999999999 3.3856449999999998 Positive [C] 2.6666669999999999 1.7049430000000001 4.1708790000000002 Positive [C] 1.236715 0.90096699999999996 1.6975800000000001 Positive [C] 1.3544970000000001 0.96319399999999999 1.904771 Negative [C] 0.18302399999999999 2.6772000000000001E-2 1.2512080000000001 Negative [C] 0.17121600000000001 2.5155E-2 1.165392 Negative [C] 0 0 NaN Negative [C] 0 0 NaN Negative [C] 0.39506200000000002 5.3231000000000001E-2 2.9319839999999999 Negative [C] 0.32323200000000002 4.5311999999999998E-2 2.3057669999999999 Positive [W] 0.3 0.178699 0.50364100000000001 Positive [W] 0.31578899999999999 0.18825900000000001 0.52971100000000004 Positive [W] 0.64285700000000001 0.35071400000000003 1.1783539999999999 Positive [W] 0.75 0.40453699999999998 1.3904780000000001 Positive [W] 0.34782600000000002 0.184888 0.65435900000000002 Positive [W] 0.38095200000000001 0.20268600000000001 0.71600600000000003 Negative [W] 3.4483E-2 5.0090000000000004E-3 0.237396 Negative [W] 3.2258000000000002E-2 4.6769999999999997E-3 0.22248000000000001 Negative [W] 0 0 NaN Negative [W] 0 0 NaN Negative [W] 0.111111 1.6934999999999999E-2 0.72901099999999996 Negative [W] 9.0909000000000004E-2 1.3715E-2 0.60258400000000001 Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Sensitivity 0.92307700000000004 0.62087999999999999 0.99597100000000005 Specificity 0.39130399999999999 0.278312 0.51646199999999998 Specificity 0.42029 0.30446299999999998 0.54503199999999996 Specificity 0.44927499999999998 0.33099699999999999 0.57322499999999998 PPV 0.222222 0.12481200000000001 0.35946400000000001 PPV 0.230769 0.129806 0.37173600000000001 PPV 0.24 0.135217 0.38486599999999999 NPV 0.96428599999999998 0.79761000000000004 0.99813300000000005 NPV 0.96666700000000005 0.80947000000000002 0.99825799999999998 NPV 0.96875 0.82001900000000005 0.998367 Pre/High sensitivity Pre/standard and low Post/high and standard Post/Low Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Estimated 95% Confidence Interval Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Value Lower Limit Upper Limit Sensitivity 1 0.62881100000000001 1 Sensitivity 1 0.62881100000000001 1 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Sensitivity 0.88888900000000004 0.50670300000000001 0.99417199999999994 Specificity 0.5625 0.37880399999999997 0.73164899999999999 Specificity 0.625 0.437496 0.783358 Specificity 0.28125 0.143986 0.46979599999999999 Specificity 0.34375 0.19173100000000001 0.53225199999999995 PPV 0.39130399999999999 0.204677 0.61217500000000002 PPV 0.42857099999999998 0.225908 0.65561000000000003 PPV 0.25806499999999999 0.12536900000000001 0.44929599999999997 PPV 0.275862 0.134459 0.47485100000000002 NPV 1 0.78124400000000005 1 NPV 1 0.79954700000000001 1 NPV 0.9 0.54115500000000005 0.99475800000000003 NPV 0.91666700000000001 0.59753900000000004 0.99563500000000005

Key Takeaways þ Rapid and continuous innovation to improve patient care þ Utilization of cutting edge science and technologies. þ Multiple proprietary technologies þ Extensive menu of multi-modality tumor profiles þ Leading provider of Liquid Biopsies

Questions and Answers

This Concludes our Investor Day Presentations. Thank you for joining us. Should you have any questions on any of these materials, please contact: Steven Jones Executive Vice President & Director of Investor Relations (239) 325-2001 steven.jones@NeoGenomics.com Thank you for joining us.